Abstract
When building genetic maps, it is necessary to choose from several marker ordering algorithms and criteria, and the choice is not always simple. In this study, we evaluate the efficiency of algorithms try (TRY), seriation (SER), rapid chain delineation (RCD), recombination counting and ordering (RECORD) and unidirectional growth (UG), as well as the criteria PARF (product of adjacent recombination fractions), SARF (sum of adjacent recombination fractions), SALOD (sum of adjacent LOD scores) and LHMC (likelihood through hidden Markov chains), used with the RIPPLE algorithm for error verification, in the construction of genetic linkage maps. A linkage map of a hypothetical diploid and monoecious plant species was simulated containing one linkage group and 21 markers with fixed distance of 3 cM between them. In all, 700 F2 populations were randomly simulated with 100 and 400 individuals with different combinations of dominant and co-dominant markers, as well as 10 and 20% of missing data. The simulations showed that, in the presence of co-dominant markers only, any combination of algorithm and criteria may be used, even for a reduced population size. In the case of a smaller proportion of dominant markers, any of the algorithms and criteria (except SALOD) investigated may be used. In the presence of high proportions of dominant markers and smaller samples (around 100), the probability of repulsion linkage increases between them and, in this case, use of the algorithms TRY and SER associated to RIPPLE with criterion LHMC would provide better results.
Similar content being viewed by others
Introduction
The construction of accurate genetic linkage maps is of crucial importance in genetic studies. The study of genome evolution processes (Paterson et al., 2000), phylogeny (Ahn and Tanksley, 1993; Bowers et al., 2003) and mapping of quantitative trait loci (Lander and Botstein, 1989; Zeng, 1993) are amongst its applications. The maps are basically built in a two-step procedure: (i) assigning markers into linkage groups and (ii) ordering the markers within the groups (Wu et al., 2003).
The ordering of markers within linkage groups is considered a special case of the classical traveling salesman problem (Doerge, 1996; Liu, 1998; Mester et al., 2003; Tan and Fu, 2006). Basically, the problem consists in choosing the best order among m!/2 possible orders (m=number of markers). When m gets larger, the number of orders is unwieldy. For example, for m=15, there are ∼6.54 × 1011 orders to be analyzed, which is not feasible with the current computational power available. Therefore, approximate solutions that allow large-scale genetic mapping have been proposed (Liu, 1998).
Several algorithms to obtain approximate solutions for ordering markers have been proposed, including simulated annealing (Thompson, 1984; Weeks and Lange, 1987), stepwise likelihood (Lathrop et al., 1984), branch and bound (Lathrop et al., 1985), try and ripple (Lander et al., 1987), seriation (SER) (Buetow and Chakravarti, 1987), rapid chain delineation (RCD) (Doerge, 1996), genetic and evolutionary algorithm (Mester et al., 2003), recombination counting and ordering (RECORD) (Van Os et al., 2005) and unidirectional growth (UG) (Tan and Fu, 2006). Some of them have been implemented into friendly user softwares, such as Linkage (Lathrop et al., 1984), MAPMAKER/EXP (Lander et al., 1987), JoinMap (Stam, 1993), Emap from the program suite QTL Cartographer (Basten et al., 2003) and OneMap (Margarido et al., 2007). Moreover, there are several criteria to evaluate and compare the orders, such as, sum of adjacent recombination fractions (SARF, Falk, 1989), product of adjacent recombination fractions (PARF, Wilson, 1988), sum of adjacent LOD scores (SALOD, Weeks and Lange, 1987), SALOD-equivalent number of fully informative meiosis (SALEQ, Edwards, 1971; Olson and Boehnke, 1990), SALOD-polymorphism information content (SALPIC, Botstein et al., 1980; Olson and Boehnke, 1990), likelihood through hidden Markov chains (LHMC, Lander and Green, 1987), least squares (Weeks and Lange, 1987) and weighted least squares (Stam, 1993).
When building genetic maps, it is necessary to choose from several algorithms and criteria, and the choice is not always simple. Using simulations, Olson and Boehnke (1990) have compared eight criteria to order a moderate number of markers (m=6). Wu et al. (2003) have evaluated the efficiency of the criteria SARF, PARF, SALOD and LHMC, along with the algorithm SER in situations with m=5. Hackett and Broadfoot (2003) have compared the efficiency of the criteria weighted least squares, SARF and LHMC in maps with 10 loci. In general, it is noticeable that none of the aforementioned works have considered the ordering problem in highly saturated maps, which is currently a very common situation due to the great availability of molecular markers. Moreover, those studies have not taken into account F2 populations and missing data in saturated maps. The presence of heterozygous loci in these populations may impair the construction of maps with the occurrence of dominant markers and missing data (Jiang and Zeng, 1997).
In this context, the goal of the present work was to evaluate the efficiency of the algorithms TRY, SER, RCD, RECORD and UG and of the criteria PARF, SARF, SALOD and LHMC used along with the algorithm RIPPLE. Situations currently found in genetic mapping and not covered in the previous studies, such as saturated maps, F2 populations, dominant markers and missing data were considered. This approach aims to provide geneticists with practical orientations to correctly choose the best combination of method and criterion when building genetic maps.
Materials and methods
Simulations
A linkage group from a hypothetical monoecious plant species, with 21 molecular markers separated by a fixed distance of 3 cM, was considered. Four sequence patterns for molecular markers were simulated: (i) CCCC; (ii) CDCD; (iii) CDDD and (iv) CDDRD (C: co-dominant marker; D and DR: dominant marker in coupling and repulsion phase, respectively, with dominant alleles from distinct parents). Each pattern was sequentially repeated five times and supplemented with a co-dominant marker at the final position, totalizing 21 markers. It is assumed that these four situations represent saturated kernels that will be ordered within real maps with many markers. As the ordering is carried out within each linkage group, it was not necessary to stipulate a chromosome number for the species, since the procedures presented here would be repeated. For each situation, F2 populations of n=100 and n=400 originated from homozygous lines were simulated, according to the scheme presented by Basten et al. (2003). Similarly, missing data were simulated by random removal of 10 and 20% of the data. In total, 24 types of experimental populations were simulated (2 sample sizes × 4 marker patterns × 3 amounts of missing data: 0, 10 and 20%). To analyze the algorithms and criteria, the Monte Carlo method was used, repeating the simulations 700 times, with each simulation being considered a replicate (Manly, 1997). Algorithms and criteria were applied to each of the 16 800 simulated samples.
Pair-wise estimations
As some algorithms use estimates of the recombination fractions and their respective LOD scores, those values were obtained for all pair-wise combinations of the markers, represented by matrices R=[r̂MiMj]21 × 21 and LOD=[lodMiMj]21 × 21, where r̂MiMj is the maximum likelihood estimate of the recombination fraction between the markers Mi and Mj (for Mi=Mj, r̂MiMj=0) and lodMiMj is the respective LOD score (Liu, 1998). Because of the fact that F2 populations were analyzed, the algorithm EM (Dempster et al., 1977; Liu, 1998; Lange, 2002) was used to obtain r̂MiMj. The estimated recombination fractions between markers were converted to distances using Haldane's (1919) mapping function.
Ordering algorithms
TRY (Lander et al., 1987): initially, markers were randomly taken and ordered through hidden Markov chain (Lander and Green, 1987) through exhaustive search (evaluation of 5!/2=60 possible orders). In the following step, a new marker was positioned at the beginning, in the end and in the middle of the m′ ordered markers, verifying which position corresponded to the highest likelihood. This step was repeated until all the markers were positioned.
SER (Buetow and Chakravarti, 1987): the map was initiated with each one of the m=21 markers and the matrix R. Considering Mi as the initial marker, Mj was positioned to the right of Mi if the recombination fraction between them was the smallest fraction between Mi and the other m−1 markers. From the remaining m−1 markers, Mk was chosen if it had the smallest recombination fraction with Mi. The recombination fractions of Mk and both external loci to the positioned markers, Mleft (the most external marker to the left) and Mright (the most external marker to the right) were compared. If r̂MkMright>r̂MkMleft, Mk was positioned to the left of the group of markers, and if the relationship was inverse, to the right. In the case of ties, the internal loci of the group already positioned were considered. The procedure was repeated until all the markers were positioned, therefore providing 21 orders (one for each marker at the initial position). For each order, the continuity index was calculated CI=∑i<jr̂MiMj/(i−j)2. The best order was considered the one that gave the smallest CI value (Liu, 1998).
RCD (Doerge, 1996): the marker pair (Mi,Mj) exhibiting the smallest recombination fraction among all markers was considered. The next unmapped marker with the smallest recombination fraction to the aforementioned pair was positioned next to them. The procedure was repeated until all markers were positioned on the map.
RECORD (Van Os et al., 2005): based on the expected number of recombination events, an S matrix was constructed, S=[SMiMj]21 × 21 (for Mi=Mj, SMiMj=0). The procedure to obtain S is based on the expected number of crossovers between marker pairs, conditioned by the observation of the markers’ phenotype. The optimization criterion COUNT for a sequence of m markers may be calculated by COUNT=∑i=1m−1SMiMi+1, where smaller COUNT values correspond to better orders. Map building was carried out by randomly taking two markers and positioning a third one at the beginning, at the end and between them. The marker was fixed at the position that gave a smaller value of COUNT. Similarly, the remaining markers were positioned at pre-established orders until completion of the map. Subsequently, a search for smaller values of COUNT was performed, inverting the position on the map of subsequences of size m′=2, … , 20. If the map resulting from the inverted positions presented a COUNT value smaller than the previous one, it was kept. The procedure was repeated 10 times and the sequence presenting smaller COUNT value was chosen.
UG (Tan and Fu, 2006): based on the R matrix, the distance between all loci was calculated by dij=r̂ij+(2/nij)∑kr̂ikr̂jk, for every k, with r̂ij>r̂ik, r̂ij>r̂jk, and nij individuals. The value Tij=2dij−(∑k≠idik+∑k≠jdjk) was calculated for every i<j. The terminal end of the map was defined by taking the pair of markers (f, g) that presented the smallest value of T. The pair (f, g) was then designed locus m+1 and its distance to the remaining markers was determined by dim+1=1/2(dif+dig−dfg) if (dif+dig)>dfg, if not, dim+1=0. The calculation Wim+1=(m−2)dim+1−∑k≠idik was also performed and the locus that minimized the value Wim+1 (called locus h) was placed on the map. The partial map resultant map was f-g-h if dfh>dgh or h-f-g if otherwise. Considering k=2, the partial distance of the map with the remaining markers was updated: dim+k=min(dim+k−1,dij). The value Wim+k=(m−k−1)dim+k−∑k≠idik was calculated and the locus that minimized W was added to the map. The last two steps were repeated, taking k=3, … , 20 to obtain the complete map.
The algorithm RIPPLE and criteria to evaluate the orders
The algorithm RIPPLE (Lander et al., 1987) allows verification of mistakes in the marker ordering and was used in all maps built earlier. It was done by permutation of a window of m′ markers (m′<m) and comparison of the m′!/2 resulting maps. Initially, positions 1, …, m′ were permutated, then positions 2, … , m′+1 and so on until the whole map was covered. The criteria SARF= (Falk, 1989), PARF= (Wilson, 1988) and SALOD= (Weeks and Lange, 1987) were used to evaluate the orders obtained after these permutations. The orders that exhibited smaller values of SARF and PARF and higher values of SALOD were considered more likely to be correct. In the present case, m′=6 was used.
Moreover, the criterion LHMC (Lander and Green, 1987) was used, by which the multipoint estimates of recombination fractions were calculated and the likelihood of the orders was compared. In this case, the window size was of m′=4, as higher numbers of orders impair the performance of Monte Carlo analyses for LHMC, because its calculation depends on intensive computational power.
All the simulated populations had maps constructed using five algorithms and four criteria (20 cases), considering also maps without the use of RIPPLE (five cases). Therefore, 420 000 maps were obtained (16800 populations × 25 combinations of algorithms and criteria). All simulations, algorithms and criteria were implemented with codes written into the R software (R Development Core Team, 2008). To calculate the criterion LHMC, source-codes in C language of the function est.map, and part of the free software qtl (Broman et al., 2008) were also used.
Evaluation of the results
To evaluate the orders provided by the algorithms and criteria, the absolute value of the Spearman's rank correlation coefficient ρ (Spearman, 1904) was calculated according to the formula ρ=1−{6∑i=1mdi2/m(m2−1)}, where di is the difference between the rank of marker Mi on the order obtained from a given procedure, and the rank of marker Mi on the actual order (simulated). The estimated distances between the markers were compared with the actual distances by calculating the average Euclidean distance D between the distances of the estimated map and the distances from the actual map: D=[(m−1)−1(d̂−d)′(d̂−d)]1/2, where d̂ is the vector of estimated distances for a given map, d is the vector for the actual distances, and ′ indicates vector transposition. Therefore, a value of 1 cM of average Euclidean distance indicates that the considered maps differ with an average of 1 cM between each other.
Results
Order of the markers
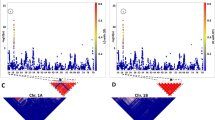
The distribution of the correlation coefficients between the order of the estimated map and the actual map showed that, in general, in the absence of dominant markers in repulsion phase (patterns CCCC, CDCD, CDDD) all the algorithms presented high correlations and small inter-quarterly amplitudes for n=100 and n=400 (Figure 1). For n=100, the algorithms RECORD and SER were slightly better than the others. Only 2.8% of the correlations were smaller than 0.95 for the algorithm RECORD and 3.0% for the algorithm SER. For the algorithms TRY, UG and RCD, the values were of 4.2, 4.8 and 7.7%, respectively. However, the correlations smaller than 0.95 varied differently among the tested algorithms, and UG presented a higher number of values close to zero. In the presence of dominant markers in repulsion phase (pattern CDDRD), it was more difficult to obtain good maps and, in general, the algorithms TRY and SER exhibited better results, especially for n=100. In these cases, best results were obtained in the absence of RIPPLE, or using RIPPLE with LHMC criterion. For n=400, the algorithms RECORD and UG also exhibited adequate results, although slightly inferior to those obtained with TRY and SER. It was also observed for the patterns CDCD, CDDD and CDDRD that the algorithm RIPPLE with the criterion SALOD exhibited lower performance in comparison to the others.
Boxplots of the distribution of the correlation coefficients between the estimated map for each Monte Carlo sample and the actual map with the five evaluated algorithms: TRY, SER, RCD, RECORD and UG. Samples with n=100 (a) and n=400 (b) individuals. On the horizontal axis, the colors indicate the pattern of the simulated markers, the amount of missing data and the criterion used with the algorithm RIPPLE. Acronyms: see Materials and methods.
The percentage of correct orders can also be used to evaluate the algorithms (Table 1). However, these results should be interpreted carefully, as inversions in closely linked markers cause little problems in practical situations, but can reduce the number of correct orders. In the absence of RIPPLE, the algorithms TRY and RECORD provided higher rates of correct orders than the others. For example, for n=100, the averages of correct orders for TRY and RECORD were 39.5 and 38.2%, respectively, and 11.9, 26.1 and 27.5% for SER, RCD and UG, respectively. However, using RIPPLE with the criteria SARF and PARF, the advantage was decreased and all the tested algorithms showed similar percentages of correct map orders. Similarly, it was observed that the use of SALOD resulted in significantly lower frequencies of correct orders in all situations involving dominant markers.
In general, the percentage of correct orders for LHMC was intermediate between those from PARF and SARF and the criterion SALOD. It was also observed that the algorithm TRY is less influenced by missing data, showing a smaller decrease in the number of correct orders in comparison to the remaining algorithms with increasing amounts of missing data. For n=400, all the algorithms presented good results, except for CDDRD.
The use of algorithm RIPPLE and criterion SALOD with patterns CDDD and CDCD (Table 1) exhibited results inferior to the other situations. For the first, the percentage of correct orders ranged from 9.7 to 1.3% (n=100) and 17.6 and 15% (n=400). For the latter, the percentage was zero for all the tested algorithms with both population sizes. For the pattern CDDRD and n=100, the algorithm TRY was slightly superior to the others, reaching 11.1% of correct orders when associated to algorithm RIPPLE and criterion LHMC in the absence of missing data. For n=400, algorithm TRY also exhibited superior results in comparison to the other, with the percentage of correct orders ranging from 18.0% (SARF with 20% of missing data) to 76.6% (LHMC without missing data). Algorithm RCD presented inadequate results for the aforementioned pattern of markers, even when combined with RIPPLE and several criteria for error verification. The algorithm UG was superior to RCD but slightly inferior to the others.
Distance between markers
In general, estimates of the distances between markers in all simulations studied showed good precision. For patterns CCCC, CDCD and CDDD, the average of the Euclidean distances between the distances on the estimated maps and on the actual maps were close to zero. For n=100, they ranged from 0.39 to 0.71 cM for the algorithm TRY, from 0.29 to 0.64 cM for SER, from 0.29 to 0.66 cM for RCD, from 0.29 to 0.64 cM for RECORD and from 0.29 to 0.72 cM for UG. For n=400, the distances were very similar for all the algorithms, with the averages ranging from 0.14 to 0.30 cM (values not showed on Figures or Tables).
As expected, for the pattern CDDRD, the averages and their confidence intervals were larger (Figure 2). For n=100, all tested algorithms showed similar results with a slight advantage towards algorithm TRY. The averages of the distances ranged from 0.50 to 1.60 cM for algorithm TRY, from 0.70 to 1.81 cM for SER, from 0.94 to 2.45 cM for RCD, from 0.78 to 2.09 cM for RECORD and from 0.83 to 2.10 cM for UG. In general, it was observed that algorithm RIPPLE used with the criteria LHMC and SALOD exhibited smaller confidence intervals. In the case of algorithm TRY, the distances obtained without the employment of RIPPLE were as adequate as those obtained using algorithm RIPPLE and LHMC. For n=400, the averages and the amplitudes of the Euclidean distances were smaller. The average of the distances with algorithm TRY ranged from 0.25 to 0.84, with SER from 0.31 to 0.89, with RCD from 0.36 to 1.28, with RECORD from 0.32 to 0.88 and with UG from 0.32 to 0.96. For algorithm UG, it was observed that the criteria PARF and SARF used with RIPPLE presented inferior results than those obtained with the criterion LHMC. It was also verified that the missing data had little influence on the calculation of the distances.
Average Euclidean distances between actual distances and the distances estimated for pattern CDDRD with algorithms TRY, SER, RCD, RECORD and UG. The averages of the calculated Euclidean distances are represented by full dots and the amplitude between the 5th and 95th percentiles (confidence interval) is represented by lines. Acronyms: see Materials and methods.
Discussion
The simulations performed here represent saturated kernels that may be present within linkage groups in saturated maps. As the analyses are independently repeated for each linkage group, the procedures may be used for any diploid species with any number of chromosomes. Moreover, the results could be also extended to other populations, such as double haploids, backcrosses and RILs, because those are easier to analyze, as they do not have mixture of genotypic classes, which causes the main difficulties for data analysis.
It can be stated that an ideal method for map construction must provide estimated maps with correlation coefficients with the actual map close to 1, high percentages of correct orders and average Euclidean distances close to zero. It has been observed in the results that, although in many cases the correlation values are high and the distances are close to zero, the algorithms provide low percentages of correct orders in several situations. This indicates that the algorithms provided adequate orders (high correlation to the actual map), although with inversions in the order of neighboring markers, which implicates in the reduction of the number of correct orders. As explained before, these inversions cause few problems, meaning that the low number of correct orders does not necessarily mean low performance. Therefore, based on these criteria, the algorithm TRY exhibited better performance. As observed by Lander et al. (1987), multipoint estimates of the recombination fractions, as used by algorithm TRY, are preferable to two point estimates, as the first is able to bypass the error propagation due to the lack of information on some marker combinations. This advantage has been shown to be particularly important in the presence of dominant markers in repulsion, as well as in the presence of missing data. In these situations, hidden Markov chain allows the most informative markers to surpass the lack of information for the others due to their linkage, therefore providing adequate results. Distance estimates present no further problems for all algorithms, although multipoint values were slightly superior.
The main problem faced by the use of multipoint approach through hidden Markov chain and likelihood (Lander and Green, 1987) concerns the time for computational processing. This approach uses an algorithm that scales linearly (not exponentially) with the number of loci, but it may impair the build of highly saturated maps. This fact has prompted the development of novel algorithms such as SER, RCD, RECORD and UG, which were proposed to deal with the high number of markers that is becoming available. SER, which is conceptually simple and computationally easy, is based on matrices with two point estimates of recombination fractions. RCD and UG are alternative approaches to SER, constructing the maps sequentially. REC uses an algorithm that allows the ordering of groups with >500 loci.
According to Thompson (1984), a heuristic estimation or inference criterion is never as adequate as those based on likelihood; however, the easiness of computational power may prompt it as a good option. The results obtained here show that in the most difficult cases of ordering, that is, those containing dominant markers in repulsion, the best alternatives to TRY were the algorithms SER and RECORD (for larger samples). With algorithm SER, the criterion used to evaluate the orders is the continuity index from the recombination fraction matrix, that takes into account the whole matrix and not only the information for each recombination fraction separately, as for algorithms RCD, RECORD and UG. That is possibly the reason of the high performance of this algorithm even in complex situations. This is in accordance with previous studies based on double haploid populations. Hackett and Broadfoot (2003) (n=150) noticed that likelihood performed better, and Wu et al. (2003) (n=100, 150, and 200) were the algorithm SER proved to be an excellent alternative.
Wu et al. (2003) have discussed that the estimates of the recombination fractions obtained between dominant markers in repulsion in an F2 population are biased, thus suggests the use of co-dominant markers to overcome the problem. The results have shown that the combination of dominant and co-dominant markers may be a good alternative for map saturation, confirming what was pointed out by Jiang and Zeng (1997) when hidden Markov chains are used. Usually, dominant markers, such as AFLP and DArT, allow genotyping a large number of loci with relative easiness (Hansen et al., 1999; Vuylsteke et al., 1999; Akbari et al., 2006; Mace et al., 2008). Those markers could be positioned in maps that already contain co-dominant markers to form a frame. This would surpass the lack of information from the dominant markers and the map could then be built using SER or RECORD, which are more feasible than TRY in this case.
Because of computational limitations imposed by Monte Carlo analyses, the window used for algorithm RIPPLE with the LHMC criterion was m′=4 markers, whereas with criteria SARF, PARF and SALOD it was m′=6. This may explain the superiority of the criteria PARF and SARF in comparison to LHMC in cases where dominant markers in association were present. In principle, the observed superiority was unexpected as the criterion for order evaluation with algorithm TRY is LHMC and the results of the aforementioned algorithm were superior. The observed difference is possibly due to differences in the values of m′. In practical situations, where only one map is built, it is obviously recommended that m′ should be as big as possible. Using modern computers, it is possible to use m′=7, which would certainly improve the estimates of order and distance. With m′=7, it took ∼26 h to build only one map for CDDRD pattern, n=100 and 10% of missing data in a personal computer Intel Core 2 Quad, 2.4 GHz with 4GB of RAM memory. This is unfeasible for doing evaluations based on Monte Carlo methods, but could be applied by users when only one map has to be built.
The criterion SALOD presented inadequate results for the three patterns with dominant markers (CDDRD, CDCD, CDDD), in accordance with the results obtained by Olson and Boehnke (1990). They stated that criterion SALOD is sensitive to the information content of the markers and tends to cluster the most informative ones. To correct the problem, the authors have proposed the criteria SALEQ (which weights SALOD by the number of informative meiosis) and SALPIC (which takes into account the information content of the polymorphism of each marker). However, these modified criteria could not be used here, as in the experimental populations used (F2) the individuals always present two informative meiosis and the information content of the polymorphism of each marker remains constant.
As general recommendation, the simulations have shown that, in the presence of co-dominant markers only, any combination of algorithm and criteria may be used, even for a reduced population size. In the case of a smaller proportion of dominant markers, the odds of having dominant markers linked in repulsion are small and any of the algorithms and criteria (except SALOD) investigated may be used as well. However, in the presence of high proportions of dominant markers and smaller samples (around 100), the probability of repulsion linkage increases between them. In this case, the use of the algorithms TRY and SER associated to RIPPLE with criterion LHMC would provide better results.
The codes to run all algorithms and criteria presented on this paper are being incorporated in the OneMap software (Margarido et al., 2007) and will be made available in a near future. Details can be obtained with the corresponding author.
References
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S et al. (2006). Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113: 1409–1420.
Ahn S, Tanksley SD (1993). Comparative linkage maps of the rice and maize genomes. Proc Natl Acad Sci USA 90: 7980–7984.
Basten CJ, Weir BS, Zeng ZB (2003). QTL Cartographer: Version 1.17. Department of Statistics, North Carolina State University, North Carolina: Raleigh.
Botstein D, White RL, Skolnick M, Davis RW (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314–331.
Bowers JE, Abbey C, Anderson S, Chang C, Draye X, Hoppe AH et al. (2003). A high-density genetic recombination map of sequence-tagged sites for sorghum, as a framework for comparative structural and evolutionary genomics of tropical grains and grasses. Genetics 165: 367–386.
Broman KW, Wu H, Churchill G, Sen S, Yandell B (2008). qtl: Tools for analyzing QTL experiments. R package version 1.08–56. http://www.rqtl.org.
Buetow KH, Chakravarti A (1987). Multipoint gene mapping using seriation. I. General methods. Am J Hum Genet 41: 180–188.
Dempster AP, Laird NM, Rubin DB (1977). Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc 39: 1–38.
Doerge RW (1996). Constructing genetic maps by rapid chain delineation. J Quant Trait Loci 2. Electronic publication available at http://wheat.pw.usda.gov/jag/papers96/paper696/doerge2.htm.
Edwards JH (1971). The analysis of X-linkage. Ann Hum Genet 34: 229–250.
Falk CT (1989). A simple scheme for preliminary ordering of multiple loci: application to 45 CF families. In: Elston RC, Spence MA, Hodge SE, MacCluer JW (eds). Multipoint Mapping and Linkage based upon Affected Pedigree Members. Genetic Workshop 6, Liss: New York. pp 17–22.
Hackett CA, Broadfoot LB (2003). Effects of genotyping errors, missing values and segregation distortion in molecular marker data on the construction of linkage maps. Heredity 90: 33–38.
Haldane JBS (1919). The combination of linkage values, and the calculation of distance between linked factors. J Genet 8: 299–309.
Hansen M, Kraft T, Chistiansson M, Nilsson ON (1999). Evaluation of AFLP in Beta. Theor Appl Genet 98: 845–852.
Jiang C, Zeng Z-B (1997). Mapping quantitative trait loci with dominant and missing markers in various crosses from two inbred lines. Genetica 101: 47–58.
Lander ES, Green P (1987). Construction of multilocus genetic linkage maps in human. Proc Natl Acad Sci 84: 2363–2367.
Lander ES, Green P, Abrahanson J, Barlow A, Daly MJ, Lincon SE et al. (1987). MAPMAKER: an interactive computing package for constructing primary genetic linkages of experimental and natural populations. Genomics 1: 174–181.
Lander ES, Botstein D (1989). Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199.
Lange K (2002). Mathematical and Statistical Methods for Genetic Analysis, 2nd edn. Springer: New York.
Lathrop GM, Lalouel JM, Julier C, Ott J (1984). Strategies for multilocus linkage analysis in human. Proc Natl Acad Sci 81: 3443–3446.
Lathrop GM, Lalouel JM, Julier C, Ott J (1985). Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37: 482–498.
Liu BH (1998). Statistical Genomics: Linkage, Mapping, and QTL Analysis, 2nd edn. CRC Press: Boca Raton.
Mace ES, Xia L, Jordan DR, Halloran K, Parh DK, Huttner E et al. (2008). DArT markers: diversity analyses and mapping in Sorghum bicolor. BMC Genomics 9: 26.
Manly B (1997). Randomization, Bootstrap and Monte Carlo Methods in Biology, 2nd edn. Chapman & Hall/CRC: London.
Margarido GRA, Souza AP, Garcia AAF (2007). OneMap: software for genetic mapping in outcrossing species. Hereditas 144: 78–79.
Mester D, Ronin Y, Minkov D, Nevo E, Korol A (2003). Constructing large-scale genetic maps using an evolutionary strategy algorithm. Genetics 165: 2269–2282.
Olson JM, Boehnke M (1990). Monte Carlo comparison of preliminary methods for ordering multiple genetic loci. Am J Hum Genet 47: 470–482.
Paterson AH, Bowers JE, Burow MD, Draye X, Elsik CG, Jiang CX et al. (2000). Comparative genomics of plant chromosomes. Plant Cell 12: 1523–1540.
R Development Core Team (2008). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org.
Spearman C (1904). The proof and measurement of association between two things. Am J Psychol 15: 72–101.
Stam P (1993). Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J 3: 739–744.
Tan Y, Fu Y (2006). A novel method for estimating linkage maps. Genetics 173: 2383–2390.
Thompson EA (1984). Information gain in joint linkage analysis. IMA J Math Appl Med Biol 1: 31–49.
Van Os H, Stam P, Visser RGF, Van Eck HJ (2005). RECORD: a novel method for ordering loci on a genetic linkage map. Theor Appl Genet 112: 30–40.
Vuylsteke M, Mank R, Antonise R, Bastiaans E, Senior ML, Stuber CW et al. (1999). Two high-density AFLP linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor Appl Genet 99: 921–925.
Weeks D, Lange K (1987). Preliminary ranking procedure for multilocus ordering. Genomics 1: 236–242.
Wilson SR (1988). A major simplification in the preliminary ordering of liked loci. Genet Epidemiol 5: 75–80.
Wu J, Jenkins J, Zhu J, Mccarty J, Watson C (2003). Monte Carlo simulations on marker grouping and ordering. Theor Appl Genet 107: 568–573.
Zeng Z-B (1993). Theoretical basis of separation of multiple linked gene effects on mapping quantitative trait loci. Proc Natl Acad Sci USA 90: 10972–10976.
Acknowledgements
M Mollinari was partially supported by Conselho Nacional de Desenvolvimento Tecnológico (CNPq—grant 132314/2006–1) and by Fundação de Amparo à Pesquisa de São Paulo (FAPESP—grant 2008/54402–4). GRA Margarido is recipient of research fellowship from FAPESP (grant 2007/02775–9). AAF Garcia is recipient of research fellowship from CNPq (grant 308139/2007–0). The authors thank Centro Nacional de Processamento de Alto Desempenho (CENAPAD-SP) for computational resources.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mollinari, M., Margarido, G., Vencovsky, R. et al. Evaluation of algorithms used to order markers on genetic maps. Heredity 103, 494–502 (2009). https://doi.org/10.1038/hdy.2009.96
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2009.96
Keywords
This article is cited by
-
QTL detection for growth and latex production in a full-sib rubber tree population cultivated under suboptimal climate conditions
BMC Plant Biology (2018)
-
SSR-enriched genetic linkage maps of bermudagrass (Cynodon dactylon × transvaalensis), and their comparison with allied plant genomes
Theoretical and Applied Genetics (2017)
-
The Sweet Passion Fruit (Passiflora alata) Crop: Genetic and Phenotypic Parameter Estimates and QTL Mapping for Fruit Traits
Tropical Plant Biology (2017)
-
High-resolution mapping and new marker development for adult plant stripe rust resistance QTL in the wheat cultivar Kariega
Molecular Breeding (2014)
-
A novel linkage map of sugarcane with evidence for clustering of retrotransposon-based markers
BMC Genetics (2012)