Abstract
The spatial genetic structure of the European ground squirrel, a species characteristic of the short-grass steppe, was investigated on the basis of a 1140-bp cyt b gene sequence. The phylogeographical architecture of this species is expected to shed light on the putative long-term presence of the steppic ecosystem in south-eastern Europe and the evolutionary consequences of glacial cycles as forcing factors in speciation. Among 31 haplotypes, three highly divergent phylogenetic lineages (Southern, Northern and Jakupica) were recognized. This result suggests the past existence of an allopatric fragmentation event caused by effective biogeographical barriers. The Southern lineage consisted of the southernmost populations, those from Greece, Macedonia and European Turkey, and showed the highest divergence from all other samples. Haplotypes of the Northern lineage showed little geographical structure, with dispersal on both sides of the Danube River and in both of the two main geographical fragments of the species. The Jakupica lineage is a geographical isolate on a high plateau in central Macedonia. The estimated time for divergence of the Southern lineage (ca. 0.58 Mya) suggests the long-term persistence of a short-grass steppic refugium in the southern Balkans. Although the divergence between the Northern and Jakupica lineages occurred more recently (ca. 0.3 Mya), it still putatively predates two glacial cycles. The three phylogeographical lineages of the European ground squirrel should be regarded as independent units for conservation management purposes.
Similar content being viewed by others
Introduction
In Europe, most of the last 2 million years have consisted of glaciations, punctuated by warmer but relatively short interglacial episodes. The biogeographical dynamics of repeated range contractions and expansions have left footprints in the genomes of species and populations (Hewitt, 2000). The evolutionary consequences of glacial cycles as forcing factors in speciation, as well as the timing of the origin of modern species, both continue to be topics of discussion and disagreement. Although some studies suggest that the entire Pleistocene, including the last two glacial cycles, was important for speciation (Johnson and Cicero, 2004; Lister, 2004), others claim that speciation and extinction rates remained constant and that speciation events extended over the past 5 Myr (Zink et al., 2004). In either case, many taxa began to diverge during glacial–interglacial cycles, when they were trapped in relatively small refugial areas, but complete speciation had not yet occurred (Hewitt, 2000). Most phylogeographical studies have shown that Southern Europe maintained major glacial refugia throughout the entire last 2 million years, although some species may also have survived in refugia to the north and east (for example, Bilton et al., 1998; Kotlík et al., 2006). At the end of the Last Glacial Maximum, some refugial populations with divergent genomes remained restricted to their former glacial refugia, whereas others expanded (for example, Hewitt, 2000). Phylogeographical studies are available for a considerable number of temperate woodland species that persisted within southern refugia (Hewitt, 2000). In contrast, much less is known about the glacial histories of steppic species that thrived during glacial periods in areas north of the southern refugia, where the environment was adverse for the majority of temperate species (Neumann et al., 2005).
Natural grasslands (steppes) are a common biome in areas with climates that are too dry or cold to develop climax woodland. Although this ecosystem is widespread in the Palearctic, it is sparse in Europe, anywhere west of Russia. Thus, a limited number of model species are available when studying the impact of glacial–interglacial cycles on steppic taxa at the western border of their current distribution. Moreover, the majority of these model species are restricted to the lowlands of the Carpathian basin and the topographically diverse Balkan Peninsula. Given the complexity of the region, the existence of refugia for steppic mammals within the Balkans appears a likely, yet largely untested, scenario. Chromosomal divergence within the lesser mole rat, Spalax leucodon (Savi and Soldatovi, 1984), supports this theory, but modern phylogeographical studies have only minimally tested this hypothesis.
Here, we focus on the European ground squirrel (Spermophilus citellus). Although this rodent can tolerate a wide range of abiotic conditions, it is strictly tied to short-grass steppe environments. This medium-sized hibernator is widespread, ranging from the semi-arid Mediterranean and Black Sea coasts up to alpine pastures at >2000 m altitude (Ružić, 1978). The European ground squirrel is the westernmost representative of ground squirrels and has seemingly been confined to its current range since its earliest appearances in fossil records (Kowalski, 2001). The current range for S. citellus is in two main fragments, separated by the Carpathian Mountains, with additional small isolates along its southern and eastern margin. At a smaller scale, this range is further fragmented along an altitudinal gradient, and high mountain populations are frequently isolated (Ružić, 1978). In the Balkan Peninsula, which forms a significant portion of the species' range, nonmetric cranial variation has revealed significant interpopulation divergence in southern fragments, suggesting their isolation over a substantial time period (Kryštufek, 1990).
The European ground squirrel seems to be a suitable model species for testing the hypothesis on the existence of refugia for steppic taxa within the Balkans because of its fragmented range, significant intraspecific variation, high habitat specificity (Ružić, 1978) and the evidence of deep divergence among haplotypes from the eastern portion of its range (Gündüz et al., 2007). In establishing the first cytochrome b (cyt b) phylogeographical study of a steppic mammal in the Balkans, we wanted to answer three questions. First, did S. citellus survive glacial–interglacial cycles in several glacial refugia? Second, which parts of the highly geomorphologically diverse landscape of southeast Europe acted as the principal Pleistocene refugia for this specific steppic taxon? Finally, do the DNA footprints of a steppic species suggest the long-term persistence of a short-grass steppe in the Balkans? This final question addresses the existence of stable glacial refugia in this region. Considering the limitations of reconstructing past environments on the basis of pollen analysis (Rackham, 1998), the phylogeographical architecture of a characteristic steppic species has the potential to provide an alternative test of the environmental history of a region.
Materials and methods
Sampling
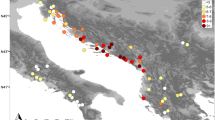
This study consisted of 26 S. citellus specimens from 11 locations ranging from the Czech Republic to European Turkey and beyond. Eleven cyt b sequences were downloaded from GenBank (Table 1, Figure 1).
Tentative range of S. citellus (shaded) with location of analyzed samples and haplotype distributions. For haplotype identities, see Table 1.
DNA extraction, PCR amplification and sequencing
A 2 × 2 mm sample was cut from tissue preserved in ethanol and air-dried in sterile conditions. DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). A PCR was carried out in a total volume of 25 μl containing 3.0 mM MgCl2, 0.3 μM of forward and reverse primers (Harrison et al., 2003), 0.2 mM dNTPs and 1 unit of Taq polymerase (Fermentas, Hanover, MA, USA) in the supplied buffer, which contained (NH4)2SO4. Cycling conditions included an initial denaturation step at 94 °C for 2 min, followed by 40 cycles of the following: denaturation (30 s at 96 °C), primer annealing (1 min at 52 °C) and extension (2 min at 72 °C). Forward and reverse sequencing was carried out on the ABI PRISM 3130 Genetic Analyzer (Applied Biosystems) using chemistry from BigDye Terminators (Applied Biosystems, Foster City, CA, USA).
Sequence analyses
The program CodonCode Aligner 1.63. (Ewing et al., 1998) was used to align forward and reverse sequences. The resulting consensus sequences for each individual were aligned using ClustalW 4.0 (Thompson et al., 1997), implemented in the MEGA package 4.0 (Tamura et al., 2007). This implementation was accomplished in combination with Bioedit 7.09 (Hall, 2004). Nucleotide and amino acid compositions were analyzed using the MEGA program. The total number of base frequencies at each position was estimated using the DAMBE 4.2.13 program (Xia and Xie, 2001).
Phylogenetic methods
The Akaike information criterion, hierarchical likelihood ratio test and Bayesian information criterion (implemented in the Modeltest 3.7. program, Posada and Crandall, 1998) were used to identify the most appropriate model of DNA substitution for the data. This model was subsequently used to calculate pairwise genetic distances among haplotypes. The phylogenetic relationships among haplotypes were reconstructed using two different optimality criteria: minimum evolution (neighbor joining of best model distances, NJ) and Bayesian inference of phylogeny (BI). An NJ tree implemented in PAUP 4.0b10 (Swofford, 2002) was constructed on the basis of the GTR+G+I substitution model (see below) using the estimated parameters. Branch reliability was estimated by a nonparametric bootstrap resampling with 10 000 replicates. For NJ trees, a bootstrap of 70% is often cited as the cutoff for a ‘reliable’ branch (Hillis and Bull, 1993), and that value was used here. S. taurensis, which is thought to be the closest relative to S. citellus (Gündüz et al., 2007), was used as the outgroup (accession no. AM691690).
The Bayesian analysis was carried out with the MrBayes 3.0 program (Huelsenbeck and Ronquist, 2001). Four Monte Carlo Markov chains were run simultaneously for 2 × 106 generations, with the resulting trees sampled every 100 generations. The likelihood scores reached stability after approximately 2 × 105 generations. As the first 10% of trees were discarded as ‘burn-in,’ only the last 180 000 trees were used to compute the 50%, majority rule consensus tree. Bayesian posterior probabilities (BPP) were used to assess branch support of the BI tree; we considered a BPP >0.90 as the cutoff for ‘moderate’ support (Buzan et al., 2008).
DNA net and mean divergences (Da and Dxy) were estimated under the K2P model and the substitution models, as determined in Modeltest using MEGA 4.0 (Tamura et al., 2007). Standard errors were estimated from 10 000 bootstrap replicates.
A minimum spanning network was constructed using both the MINSPNET algorithm that is available in the ARLEQUIN 3.1 program (Schneider et al., 2000) and the TCS 1.21 algorithm (Clement et al., 2000).
Geographical distribution of genetic variation
To assess whether genetic diversity differed between main lineages, nucleotide and haplotype diversities were calculated for each lineage. Nucleotide and haplotype diversities (π) within phylogenetic lineages were estimated using DnaSP 4.10.9 (Rozas et al., 2003).
Molecular clock estimates
Divergence time between groups was estimated as T=Da/2 μ, where 2 μ is the divergence rate. Ninety-five percent confidence intervals for the divergence times were calculated as ±1.96 s.e. of the net distances. In the estimation of molecular clocks, we followed Gündüz et al., 2007, who estimated divergence times in S. xanthoprymnus from cyt b data. Their divergence rate (approximately 2% net distance per Myr) estimate was based on the molecular data developed for Spermophilus by Harrison et al., 2003. This substitution rate is probably dependent on the calibration point (see Ho et al., 2005, 2007), and more data are needed before any sort of time dependence of estimated rates can be formulated and evaluated over the expected divergence times (Bandelt, 2008).
Results
Sequence data
The 42 S. citellus samples yielded 31 different cyt b haplotypes. Of the 1140 bp sequenced, 69 polymorphic sites were found. These 69 polymorphic sites included a total of 70 mutations, 43 of which were parsimony informative. No stop-codon insertions or deletions were observed in the alignment. As expected under neutral evolution (Martin and Palumbi, 1993), the majority of polymorphic sites were at third positions (49 variable sites, 71.0% of all variable sites), followed by first positions (15 variable sites, 21.7% of all variable sites) and second positions (5 variable sites, 7.3% of all variable sites).
Under all three approaches for the selection of the best substitution model in the Modeltest 3.7 program, the General Time Reversible model (GTR+G+I) was chosen as the most appropriate for the data set. The model included unequal base frequencies (A=0.291, C=0.255, G=0.121 and T=0.333), a proportion of invariable sites of 0.648 and a substitution rate among sites following a gamma distribution (α=0.869).
Phylogenetic analyses
As the phylogenetic methods (NJ and BI) gave identical arrangements of the main branches, the relationship between haplotypes is presented only for the BI analysis (Figure 2). Two major groups emerged with a mean sequence divergence of 1.97%±0.33 and a net divergence of 1.16%±0.28. These groups were well supported (BPP=0.96) and showed a strong geographical association. The first major phylogeographical lineage (the Southern lineage) consisted of samples from the very southern margin of the European ground squirrel's range in the lowlands of Turkish Thrace, Greece and Macedonia. A single haplotype from the Danube delta (Romania, as described by Gündüz et al., 2007) also belonged to this lineage. These data suggested a limited sympatry between the Southern and Northern phylogeographical groups along the eastern coast of the Black Sea. The second major phylogeographical group contained the remaining samples and was further divided into two sublineages. The Jakupica lineage is an isolate on a high plateau of Mt Jakupica in Macedonia. The Northern lineage consisted of all the remaining haplotypes (Figure 2). Support for these two lineages was strong (BPP=0.97 for BI and BP=89% for NJ), and the mean sequence divergence between them was 0.98%±0.23 (net divergence 0.06%±0.02). There is moderate support (BPP=0.94 for BI and BP=86% for NJ) for a further phylogeographical subdivision of the Northern lineage. Three haplotypes (N10, N11 and N12) are allopatric to the easternmost portion of the species' range in Romania and Moldova, whereas the remaining nine haplotypes of the Northern lineage are widely dispersed in both main segments of the species' range and on both sides of the Danube River (Figure 1).
Fifty percent majority rule consensus tree of 180 000 trees from a Bayesian analysis of the 1140-bp cyt b gene of S. citellus, rooted with S. taurensis. The branching pattern and branch lengths follow the Bayesian analysis, whereas numbers on the branches correspond to posterior probability values in the Bayesian inference (BI) (>0.90) and bootstrap support in the neighbor joining (NJ) (>70%) analyses, respectively. For abbreviations see Table 1.
As the Northern, Southern and Jakupica lineages could not be connected under the 95% confidence limits in TCS (they are separated by ⩾13 substitutions), separate networks were calculated, which resulted in the same topology as estimated in ARLEQUIN. Minimum spanning networks (Figure 3) showed similar results to BI and NJ trees. The Northern and the Southern lineages, as evidenced in our phylogenetic analysis, were similarly retrieved and separated by 21 mutational steps. In addition, the subdivision between the Northern and the Jakupica lineages (13 mutational steps) was also reflected in the minimum spanning tree. Within the Northern lineage, the three haplotypes (N10–N12) from Moldova and eastern Romania were the most divergent, separated by seven mutational steps. Relatively high numbers of mutational steps joined the haplotypes in the Southern lineage. Conversely, low divergence was seen among the five haplotypes from the Jakupica lineage. These haplotypes were separated from one another by only single steps.
An unrooted minimum spanning tree constructed for the 31 haplotypes detected in this study. The numbers of mutational steps joining the haplotypes are indicated along the connecting branches. The Southern lineage (S) is separated from the Northern lineage (N) by 21 mutational steps, whereas the Northern and Jakupica (J) lineages are separated by 13 mutational steps. The correspondence between haplotype acronyms and their geographical origins is shown in Table 1. Small dots represent hypothetical haplotypes not found in the sample.
Nucleotide diversity was higher in the Southern lineage (π=0.92±0.49%) than the Northern lineage (π=0.74±0.34%), although the difference was not significant (P>0.1). Haplotype diversity was very similar in the Northern (h=0.98±0.019) and Southern (h=0.97±0.025) lineages.
Divergence times
Application of the 2% divergence rate to the net divergence estimates suggests that the two main lineages separated about 0.58 Mya (95% confidence interval=0.31−0.86 Mya), whereas the divergence between the Northern and the Jakupica lineages putatively occurred by approximately 0.30 Mya (95% confidence interval=0.10−0.50 Mya).
Discussion
Phylogeography and refugia
On the basis of our mitochondrial phylogenetic analysis, S. citellus populations are divided into two main cyt b groups. The first main group (the Southern lineage) includes samples from the southern margin of the species range and one haplotype from the Danube delta in Romania. The second main group consists of two sublineages, one restricted to Mt Jakupica in central Macedonia (Jakupica lineage) and the other comprising all the remaining samples (Northern lineage). Deep genealogical divergence among the three lineages and very limited geographical overlap of their haplotype distribution indicate that they originated from an allopatric fragmentation event (Avise, 2000). This event presumes effective biogeographical barriers. Despite the clear phylogeographical structuring, this interpretation requires some caution because of limited sampling and reliance on a single locus. The results from a single gene represent only a small fraction of genetic history and provide an incomplete picture of historical processes occurring in a species (Avise 2000). Further studies of nuclear markers are thus expected to provide independent tests of the phylogeographical structure that was constructed on the basis of the cyt b gene. In addition, divergence times may be overestimated because the substitution rates that were used are probably lower than the true rates (Ho et al., 2005, 2007). However, Ho et al.,'s concern regarding overestimated divergence times was recently criticized (Emerson, 2007); therefore, it is evident that further work is necessary to precisely estimate the substitution rates of cyt b sequences. We propose the possible evolutionary scenario of the European ground squirrel based on divergence times estimated using the conservative 2% substitution rate per Myr.
Considering the estimated divergence time between the two main groups (ca. 0.5 Mya), the fragmentation of the putative panmictic ancestral population occurred during the Mindel interstadial. The divergence between the Northern and the Jakupica lineages is more recent (ca. 0.3 Mya), although it still predates two main glacial cycles (Riss and Würm), and tentatively coincides with the Mindel/Riss interglacial period (Hewitt, 2000). The most plausible scenario for the evolutionary history of S. citellus since the Middle Pleistocene is thus an early vicariance of the ancestral population into two major phylogeographical lineages that remained effectively isolated afterwards. Current biogeographical barriers, the Danube River and the Carpathians, effectively fragment the recent main range of S. citellus, but were evidently not operational in the past. In the absence of an apparent biogeographical barrier between the recent ranges of the Northern and the Southern lineages, patchiness of forest and steppe habitats was likely to be the only factor isolating the two phylogroups.
Glaciations in the Mediterranean took the form of dry rather than cold periods. The landscape was typically dominated by grasses interspersed with scattered trees, resembling savannas or forest steppes, although presently, there seems to be no precisely similar environment in the world (Rackham, 1998). The pollen record from northern Greece suggests contrasting bioclimatic areas throughout the last glacial period, with certain habitats remaining immune to the extreme effects of Quaternary climate variability, preserving temperate tree populations, and other sites experiencing the pronounced aridity that favored grasses and sagebrush Artemisia (Tzedakis et al., 2002). In a manner similar to the present times, historical European ground squirrels most likely thrived in metapopulations. Metapopulation structure and dynamics were likely determined by the mosaic of forest/steppe habitat, changing in response to glacial–interglacial oscillations. Ground squirrels expanded their range as increased aridity favored grasslands; range was then reduced and fragmented in wet periods when forests expanded. Similar to recent isolates, which experienced repeated bottlenecks and are affected by inbreeding (Hulová and Sedláček, 2008), poor local survival combined with a cessation of immigration caused isolated populations to crash (Hoffmann et al., 2003). Given that S. citellus depends entirely on short-grass habitats (Ružić, 1978), grazing pressure may have also been an important factor in determining its former range throughout the glacial–interglacial cycles. However, the actual impact of grazing is unknown (Rackham, 1998). Nevertheless, the species richness of the mega-herbivorous community in the Quaternary strongly exceeded modern-day diversity throughout Europe (Guérin and Patou-Mathis 1996). This high richness suggests a higher grazing impact on grasslands.
The Northern and Southern lineages consist of deeply divergent haplotypes, the distributions of which showed only limited geographical structure. Within each major phylogeographical lineage, ground squirrel populations were presumably repeatedly fragmented when the short-grass steppe was diminished during the expanse of tall-grass steppe, shrubs and/or forests. Populations most likely survived in isolated habitats and differentiated through genetic drift or divergent selection. Such ‘micro-allopatry’ was presumably repeated during each interglacial phase and then dispersed during excessively dry glacial periods, with secondary admixture of the allopatrically evolved populations. The Jakupica lineage is clearly a marginal isolate entrapped on a high-mountain steppe island following forest expansion, most likely during the Mindel/Riss interglacial period.
The time divergences estimated between the S. citellus lineages (ca. 0.5 Mya between the Northern and Southern lineages and ca. 0.3 Mya between the Northern and Jakupica lineages) are remarkably similar to estimates of time divergence among the five phylogroups of the Anatolian S. xanthoprymnus (0.30−0.75 Mya; Gündüz et al., 2007). Given that forests covered significant portions of Asia Minor throughout the Upper Pleistocene into the early Holocene (Kosswig, 1955), the large-scale, forest-steppe patch dynamics during the glacial–interglacial cycles were possibly synchronous in the Balkans and Anatolia.
Phylogenetic relations between S. citellus and S. taurensis
Our results confirmed the monophyly of S. citellus against the newly recognized S. taurensis, a small-range species restricted to the Taurus Mountains in Anatolia (Gündüz et al., 2007). Evolutionary divergence between these two sister species was likely triggered by the submergence of the temporary Balkan–Anatolian land bridge (Kerey et al., 2004) ca. 2.5 Mya, predating the Quaternary glacial–interglacial cycles (Gündüz et al., 2007). Given that the oldest divergence in S. citellus is estimated at ca. 0.5 Mya, approximately 2 million years of evolutionary history of the European ground squirrel or its ancestors remain unknown. As the taxonomy of the Quaternary ground squirrels is unstable and rather elusive (Kowalski, 2001), paleontology is currently unable to illuminate this long evolutionary gap.
Conservation implications
Approximately half a century ago, S. citellus was considered a major agricultural pest. Subsequent transformation in farming practices, however, degraded short-grass habitats and fragmented ground squirrel populations to the extent that S. citellus is currently classified as ‘vulnerable’ in the IUCN Red List of Threatened Animals (Hulovaá and Sedláček, 2008). Our results indicate that the European ground squirrel is composed of three historically isolated, independently evolving sets of populations. The conservation of such evolutionarily significant units is regarded as an important goal in preserving within-species diversity (Moritz, 1994). Therefore, the three phylogeographical lineages of the European ground squirrel should be regarded as independent units for conservation management purposes. In particular, the two southern evolutionarily significant units (the Jakupica and Southern lineages) require imminent conservation attention. The Jakupica lineage has an extremely small and poorly documented range of occupancy (possibly <100 km2; author's unpublished data). The range of the Southern lineage, particularly its western segment, is highly fragmented, and the species' decline in Greece and Macedonia over the last few decades has been catastrophic (personal observation).
As surviving populations of the European ground squirrel are typically small, isolated or both, translocations and reintroductions were recently initiated in Central Europe as a common conservation practice (Hulovaá and Sedláček, 2008). Thus far, these measures have been restricted to populations of the Northern lineage, which we presume did not alter the phylogeographical structure of the species. Great caution is required for planning reintroductions along the southern border of the European ground squirrel's range, particularly in Bulgaria, where the ranges of the two main phylogeographical groups (Southern and Northern) are only tentatively known. This lack of knowledge increases the need for a detailed population genetic study in the area of overlap of mtDNA lineages in Bulgaria and Romania. Estimation of the degree of gene flow between the two lineages, or their possible microallopatric separation, is an area for future research.
Accession codes
Accessions
GenBank/EMBL/DDBJ
References
Avise JC (2000). Phylogeography: The History of Formation of Species. Harvard University Press: Cambridge.
Bandelt H-J (2008). Time dependency of molecular rate estimates: tempest in a teacup. Heredity 100: 1–2.
Bilton DT, Mirol PM, Mascheretti S, Fredga K, Zima J, Searle JB (1998). Mediterranean Europe as an area of endemism for small mammals rather than a source for northwards postglacial colonization. Phil Trans R Soc Lond B 265: 1219–1226.
Buzan EV, Krystufek B, Hänfling B, Hutchinson WF (2008). Mitochondrial phylogeny of Arvicolinae using comprehensive taxonomic sampling yields new insights. Biol J Linn Soc 94: 825–835.
Clement M, Posada D, Crandall K (2000). TCS: a computer program to estimate gene genealogies. Mol Ecol 9: 1657–1660.
Emerson BC (2007). Alarm bells for the molecular clock? No support for Ho et al.'s model of time-dependent molecular rate estimates. Syst Biol 56: 337–345.
Ewing B, Hillier L, Wendl MC, Green P (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185.
Guérin C, Patou-Mathis M (1996). Les grands mammifères Plio-Pléistocènes d'Europe. Masson: Paris.
Gündüz İ, Jaarola M, Tez C, Yeniyurt C, Polly PD, Searle JB (2007). Multigenic and morphometric differentiation of ground squirrels (Spermophilus, Sciuridae, Rodentia) in Turkey, with description of a new species. Mol Phylogenet Evol 43: 916–935.
Hall T (2004). Bioedit 7.0.0. North Carolina State University: Raleigh.
Harrison RG, Bogdanowicz SM, Hoffmann RS, Yensen E, Sherman PW (2003). Phylogeny and evolutionary history of the ground squirrels (Rodentia: Marmotinae). J Mamm Evol 10: 249–276.
Hewitt G (2000). The genetic legacy of the Quaternary ice ages. Nature 405: 907–913.
Hillis DM, Bull JJ (1993). An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42: 182–192.
Ho SYW, Phillips MJ, Cooper A, Drummond AJ (2005). Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol 22: 1561–1568.
Ho SYW, Kolokotronis S-O, Allaby RG (2007). Elevated substitution rates estimated from ancient DNA sequences. Biol Lett 3: 702–705.
Hoffmann I, Millesi E, Huber S, Everts LG, Dittami JP (2003). Population dynamics of European ground squirrels (Spermophilus citellus) in a suburban area. J Mammal 84: 615–626.
Huelsenbeck JP, Ronquist F (2001). MrBayes: Bayesian inference of phylogeny. Bioinformatics 17: 754–755.
Hulová š, Sedláček F (2008). Population genetic structure of the European ground squirrel in the Czech Republic. Conserv Genet 9: 615–625.
Johnson NK, Cicero C (2004). New mitochondrial DNA data affirm the importance of Pleistocene speciation in North American birds. Evolution 58: 1122–1130.
Kerey IE, Meriç E, Tunoğlu C, Kelling G, Brenner RL, Doğan AU (2004). Black sea-Marmara sea quaternary connections: new data from the Bosphorus, Istanbul, Turkey. Palaeogeogr Palaeoclimatol Palaeoecol 204: 277–295.
Kosswig C (1955). Zoogeography of the near East. Syst Zool 4: 49–73.
Kotlík P, Deffontaine V, Mascheretti S, Zima J, Michaux JR, Searle JB (2006). A northern glacial refugium for bank voles (Clethrionomys glareolus). PNAS 103: 14860–14864.
Kowalski K (2001). Pleistocene rodents of Europe. Folia Quaternaria 72: 3–389.
Kruckenhauser L, Pinsker W, Haring E, Arnold W (1999). Marmot phylogeny revisited: molecular evidence for a diphyletic origin of sociality. J Zool Syst Evol Res 37: 49–56.
Kryštufek B (1990). Nonmetric cranial variation and divergence of European sousliks (Citellus citellus) from Yugoslavia (Rodentia, Sciuridae). Boll Zool 57: 351–355.
Lister AM (2004). The impact of Quaternary ice ages on mammalian evolution. Phil Trans R Soc Lond B 359: 221–241.
Martin AP, Palumbi SR (1993). Protein evolution in different cellular environments: cytochrome b in sharks and mammals. Mol Biol Evol 10: 873–891.
Moritz C (1994). Defining evolutionarily-significant-units for conservation. TREE 9: 373–375.
Neumann K, Michaux JR, Maak S, Jansman HAH, Kayser A, Mundt G et al. (2005). Genetic spatial structure of European common hamsters (Cricetus cricetus)—a result of repeated range expansion and demographic bottlenecks. Mol Ecol 14: 1473–1483.
Posada D, Crandall KA (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818.
Rackham O (1998). Savanna in Europe. In: Kirby KJ, Watkins C (eds). The ecological history of European forest. Cab Int: New York. pp. 1–24.
Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R (2003). DNASP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Ružić A (1978). Citellus citellus (Linnaeus, 1766)—Der oder das Europäische Ziesel. In: Niethammer J, Krapp F (eds). Handbuch der Säugetiere Europas. Bd. 1, Nagetiere I. Akad Verlagsgesellschaft: Wiesbaden. pp 123–144.
Savić I, Soldatović B (1984). Karyotype evolution of the genus Nannospalax Palmer, 1903, Mammalia, in Europe. Serb Acad Sci Arts 560: 1–104.
Schneider S, Roessli D, Excoffier L (2000). ARLEQUIN, Version 2.0: A Software for Population Genetic Data Analysis. Genetics and Biometry Laboratory, University of Geneva: Geneva.
Swofford DL (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods). Version 4.0b10. Sinauer Associates, Sunderland: MA.
Tamura K, Dudley J, Nei M, Kumar S (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997). The CLUSTAL-windows interface: flexible strategies for multiple sequence aligment aided by quality analysis tools. Nucleic Acid Res 25: 4876–4882.
Tzedakis PC, Lawson IT, Frogley MR, Hewitt GM, Preece RC (2002). Buffered three population changes in a Quaternary refugium: Evolutionary implications. Science 297: 2044–2047.
Xia X, Xie Z (2001). DAMBE: Data analysis in molecular biology and evolution. J Hered 92: 371–373.
Zink RM, Klicka J, Barber BR (2004). The tempo of avian diversification during the quaternary. Phil Trans R Soc Lond B 359: 215–220.
Acknowledgements
We thank the following people who helped during fieldwork or provided tissue samples: Svetozar Petkovski (Skopje), Georg Džukić (Belgrade), Beytullah Özkan (Edirne) and Jan Matěju (Karlovy Vary). This project was supported by the Czech–Slovenian intergovernmental S&T cooperation program (project no. MEB 090802), and laboratory analyses were partially financed by the Ministry of Education of the Czech Republic (research centre no. LC06073 and project MSM 00216622416). P David Polly and six anonymous referees provided valuable comments on an earlier draft.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kryštufek, B., Bryja, J. & Bužan, E. Mitochondrial phylogeography of the European ground squirrel, Spermophilus citellus, yields evidence on refugia for steppic taxa in the southern Balkans. Heredity 103, 129–135 (2009). https://doi.org/10.1038/hdy.2009.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2009.41
Keywords
This article is cited by
-
Phylogeography of the pallid ground squirrel (Spermophilus pallidicauda Satunin, 1903) as a consequence of Quaternary changes in the Mongolian open landscape ecosystems
Mammal Research (2023)
-
Recently lost connectivity in the Western Palaearctic steppes: the case of a scarce specialist butterfly
Conservation Genetics (2020)
-
Phylogeographic patterns of steppe species in Eastern Central Europe: a review and the implications for conservation
Biodiversity and Conservation (2016)
-
An ontogenetic perspective on the study of sexual dimorphism, phylogenetic variability, and allometry of the skull of European ground squirrel, Spermophilus citellus (Linnaeus, 1766)
Zoomorphology (2013)
-
Mitochondrial cytochrome b sequences resolve the taxonomy of field mice (Apodemus) in the western Balkan refugium
Acta Theriologica (2012)