Abstract
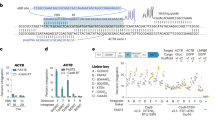
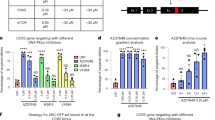
Advancements in genome editing have relied on technologies to specifically damage DNA which, in turn, stimulates DNA repair including homologous recombination (HR). As off-target concerns complicate the therapeutic translation of site-specific DNA endonucleases, an alternative strategy to stimulate gene editing based on fragile DNA was investigated. To do this, an episomal gene-editing reporter was generated by a disruptive insertion of the adeno-associated virus (AAV) inverted terminal repeat (ITR) into the egfp gene. Compared with a non-structured DNA control sequence, the ITR induced DNA damage as evidenced by increased gamma-H2AX and Mre11 foci formation. As local DNA damage stimulates HR, ITR-mediated gene editing was investigated using DNA oligonucleotides as repair substrates. The AAV ITR stimulated gene editing >1000-fold in a replication-independent manner and was not biased by the polarity of the repair oligonucleotide. Analysis of additional human DNA sequences demonstrated stimulation of gene editing to varying degrees. In particular, inverted yet not direct, Alu repeats induced gene editing, suggesting a role for DNA structure in the repair event. Collectively, the results demonstrate that inverted DNA repeats stimulate gene editing via double-strand break repair in an episomal context and allude to efficient gene editing of the human chromosome using fragile DNA sequences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Collins J, Volckaert G, Nevers P . Precise and nearly-precise excision of the symmetrical inverted repeats of Tn5; common features of recA-independent deletion events in Escherichia coli. Gene 1982; 19: 139–146.
Gordenin DA, Lobachev KS, Degtyareva NP, Malkova AL, Perkins E, Resnick MA . Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol 1993; 13: 5315–5322.
Lobachev KS, Rattray A, Narayanan V . Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front Biosci 2007; 12: 4208–4220.
DasGupta U, Weston-Hafer K, Berg DE, Local DNA . sequence control of deletion formation in Escherichia coli plasmid pBR322. Genetics 1987; 115: 41–49.
Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko PJ et al. Palindrome resolution and recombination in the mammalian germ line. Mol Cell Biol 1997; 17: 5559–5570.
Leach DR, Okely EA, Pinder DJ . Repair by recombination of DNA containing a palindromic sequence. Mol Microbiol 1997; 26: 597–606.
Eykelenboom JK, Blackwood JK, Okely E, Leach DR . SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol Cell 2008; 29: 644–651.
Farah JA, Hartsuiker E, Mizuno K, Ohta K, Smith GR . A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics 2002; 161: 461–468.
Koller BH, Smithies O . Altering genes in animals by gene targeting. Ann Rev Immunol 1992; 10: 705–730.
Capecchi MR . Altering the genome by homologous recombination. Science 1989; 244: 1288–1292.
Porteus MH, Baltimore D . Chimeric nucleases stimulate gene targeting in human cells. Science 2003; 300: 763.
Porteus MH, Cathomen T, Weitzman MD, Baltimore D . Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol 2003; 23: 3558–3565.
Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW . The double-strand-break repair model for recombination. Cell 1983; 33: 25–35.
Stuckey S, Storici F . Genetic modification stimulated by the induction of a site-specific break distant from the locus of correction in haploid and diploid yeast Saccharomyces cerevisiae. Methods Mol Biol 2014; 1114: 309–324.
Metzger MJ, McConnell-Smith A, Stoddard BL, Miller AD . Single-strand nicks induce homologous recombination with less toxicity than double-strand breaks using an AAV vector template. Nucleic Acids Res 2011; 39: 926–935.
Certo MT, Gwiazda KS, Kuhar R, Sather B, Curinga G, Mandt T et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods 2012; 9: 973–975.
Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res 2006; 34: e149.
Pattanayak V, Ramirez CL, Joung JK, Liu DR . Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods 2011; 8: 765–770.
Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol 2011; 29: 816–823.
Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 2014; 42: 7473–7485.
Mastroianni M, Watanabe K, White TB, Zhuang F, Vernon J, Matsuura M et al. Group II intron-based gene targeting reactions in eukaryotes. PLoS One 2008; 3: e3121.
Knauert MP, Glazer PM . Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum Mol Genet 2001; 10: 2243–2251.
Carroll D . A CRISPR approach to gene targeting. Mol Ther 2012; 20: 1658–1660.
Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM . DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 1998; 273: 5858–5868.
Xiao X, Xiao W, Li J, Samulski RJ . A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J Virol 1997; 71: 941–948.
Pommier Y, Leo E, Zhang H, Marchand C . DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem Biol 2010; 17: 421–433.
Stracker TH, Petrini JH . The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 2011; 12: 90–103.
Petrini JH, Stracker TH . The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol 2003; 13: 458–462.
Schwartz RA, Carson CT, Schuberth C, Weitzman MD . Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J Virol 2009; 83: 6269–6278.
Cervelli T, Palacios JA, Zentilin L, Mano M, Schwartz RA, Weitzman MD et al. Processing of recombinant AAV genomes occurs in specific nuclear structures that overlap with foci of DNA-damage-response proteins. J Cell Sci 2008; 121: 349–357.
Choi VW, McCarty DM, Samulski RJ . Host cell DNA repair pathways in adeno-associated viral genome processing. J Virol 2006; 80: 10346–10356.
Inagaki K, Ma C, Storm TA, Kay MA, Nakai H . The role of DNA-PKcs and artemis in opening viral DNA hairpin termini in various tissues in mice. J Virol 2007; 81: 11304–11321.
Hirsch ML, Storici F, Li C, Choi VW, Samulski RJ . AAV recombineering with single strand oligonucleotides. PLoS One 2009; 4: e7705.
Michel B, Grompone G, Flores MJ, Bidnenko V . Multiple pathways process stalled replication forks. Proc Natl Acad Sci USA 2004; 101: 12783–12788.
Petermann E, Helleday T . Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 2010; 11: 683–687.
Weinert T, Kaochar S, Jones H, Paek A, Clark AJ . The replication fork's five degrees of freedom, their failure and genome rearrangements. Curr Opin Cell Biol 2009; 21: 778–784.
Young Jr SM, Xiao W, Samulski RJ . Site-specific targeting of DNA plasmids to chromosome 19 using AAV cis and trans sequences. Methods Mol Biol 2000; 133: 111–126.
Hewitt FC, Samulski RJ . Creating a novel origin of replication through modulating DNA-protein interfaces. PLoS One 2010; 5: e8850.
Porteus MH, Carroll D . Gene targeting using zinc finger nucleases. Nat Biotechnol 2005; 23: 967–973.
Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA . Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J 2000; 19: 3822–3830.
Storici F, Durham CL, Gordenin DA, Resnick MA . Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc Natl Acad Sci USA 2003; 100: 14994–14999.
Storici F, Lewis LK, Resnick MA . In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol 2001; 19: 773–776.
Samulski RJ, Srivastava A, Berns KI, Muzyczka N . Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 1983; 33: 135–143.
Katz SS, Gimble FS, Storici F . To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human c ells. PLoS One 2014; 9: e88840.
Yan Z, Zak R, Zhang Y, Engelhardt JF . Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J Virol 2005; 79: 364–379.
Hirsch ML, Green L, Porteus MH, Samulski RJ, Self-complementary AAV . mediates gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Ther 2010; 17: 1175–1180.
Grieger JC, Choi VW, Samulski RJ . Production and characterization of adeno-associated viral vectors. Nat Protoc 2006; 1: 1412–1428.
Bower JJ, Karaca GF, Zhou Y, Simpson DA, Cordeiro-Stone M, Kaufmann WK . Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene 2010; 29: 4787–4799.
Acknowledgements
I thank Drs C Yin, C Hewitt, K Lobachev, and R J Samulski for molecular biology expertise, reagents and helpful discussions. This work was supported by the Northwest Genome Engineering Consortium, the Jain Foundation, and the NIH (RO1AI072176-06A1, RO1AR064369-01A1). Funding was also provided in part through a departmental Unrestricted Grant from Research to Prevent Blindness, New York, NY, USA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no conflict of interest.
Rights and permissions
About this article
Cite this article
Hirsch, M. Adeno-associated virus inverted terminal repeats stimulate gene editing. Gene Ther 22, 190–195 (2015). https://doi.org/10.1038/gt.2014.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2014.109
This article is cited by
-
BTK gene targeting by homologous recombination using a helper-dependent adenovirus/adeno-associated virus hybrid vector
Gene Therapy (2016)
-
Development of non-defective recombinant densovirus vectors for microRNA delivery in the invasive vector mosquito, Aedes albopictus
Scientific Reports (2016)
-
Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors
Nature Biotechnology (2015)