Abstract
Purpose
The craniosynostoses are characterized by premature fusion of one or more cranial sutures. The relative contribution of previously reported genes to craniosynostosis in large cohorts is unclear. Here we report on the use of a massively parallel sequencing panel in individuals with craniosynostosis without a prior molecular diagnosis.
Methods
A 20-gene panel was designed based on the genes’ association with craniosynostosis, and clinically validated through retrospective testing of an Australian and New Zealand cohort of 233 individuals with craniosynostosis in whom previous testing had not identified a causative variant within FGFR1-3 hot-spot regions or the TWIST1 gene. An additional 76 individuals were tested prospectively.
Results
Pathogenic or likely pathogenic variants in non-FGFR genes were identified in 43 individuals, with diagnostic yields of 14% and 15% in retrospective and prospective cohorts, respectively. Variants were identified most frequently in TCF12 (N = 22) and EFNB1 (N = 8), typically in individuals with nonsyndromic coronal craniosynostosis or TWIST1-negative clinically suspected Saethre–Chotzen syndrome. Clinically significant variants were also identified in ALX4, EFNA4, ERF, and FGF10.
Conclusion
These findings support the clinical utility of a massively parallel sequencing panel for craniosynostosis. TCF12 and EFNB1 should be included in genetic testing for nonsyndromic coronal craniosynostosis or clinically suspected Saethre–Chotzen syndrome.
Similar content being viewed by others
Introduction
Craniosynostoses are malformations of the developing skull characterized by the premature fusion of one or more cranial sutures. They have marked phenotypic and allelic heterogeneity and are classified into nonsyndromic and syndromic forms. Nonsyndromic craniosynostosis is an isolated finding and is classified according to the suture(s) involved. Syndromic craniosynostoses are associated with other malformations and typically have a monogenic etiology.
Early diagnosis and multidisciplinary team management play key roles in preventing clinical sequelae of untreated craniosynostosis, which can include raised intracranial pressure, visual impairment, sleep abnormalities, facial dysmorphism, and developmental delay.1 Surgical treatment of craniosynostosis is currently guided by the severity and age of onset of craniofacial abnormalities, rather than genetic etiology. However, genetic causes are more likely to require secondary craniofacial surgery or develop extracranial complications, such as requirements for hearing aids and for upper-limb surgery; a notable exception is craniosynostosis due to TCF12 variants, which has a more benign course and is less likely to require secondary craniofacial surgery.2,3,4 A molecular diagnosis can therefore reveal gene-specific prognoses for patients and guide reproductive genetic counseling.
Genetic testing for craniosynostosis in this laboratory has historically been limited to a range of hot-spot regions in FGFR1, FGFR2, FGFR3, and the single coding exon of TWIST1.1 These were the first craniofacial genes of major effect to be identified and contain gain-of-function variants associated with Crouzon, Pfeiffer, Muenke, Apert, Saethre–Chotzen, and Antley–Bixler syndromes (Table 1). This targeted approach identifies a causative variant in approximately 71–75% of probands with syndromic craniosynostosis, but rarely in those with nonsyndromic unisutural craniosynostosis.1,5,6
Additional genes have been identified more recently, associated with a range of autosomal dominant, autosomal recessive, and X-linked craniosynostoses; most notably, TCF12 (coronal), ERF (sagittal, lambdoid), FREM1 (metopic), and EFNA4 (coronal).3,7,8,9 The relative causative contributions of these genes in a clinically ascertained cohort from a multidisciplinary craniofacial clinic is unclear. Routine testing of an expanded list of genes was previously not feasible owing to considerations of cost and scale, but is now possible using massively parallel sequencing (MPS). The ability of MPS to test multiple genes simultaneously lends itself to testing in disorders that display locus heterogeneity, such as the craniosynostoses, and genes where variants are not clustered within hot-spot exons, such as TCF12.
In view of these newly identified genes, and the potential for improved diagnostic yield through MPS of multiple genes, a 20-gene custom AmpliSeq panel was designed and clinically validated on the Ion Proton sequencing instrument. Variant-negative individuals with craniosynostosis were retrospectively tested to establish analytical validity and the relative causative contribution of these craniosynostosis genes. Additional individuals with craniosynostosis were prospectively tested to verify clinical utility in an unselected referral cohort.
Materials and methods
Subjects
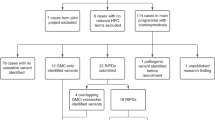
The subjects in this study included a retrospective cohort of previously undiagnosed patients (N = 233), as well as a prospective patient cohort (N = 76). Retrospective analysis used archived DNA from patients referred to a clinically accredited laboratory, who did not have a molecular diagnosis after Sanger sequencing of hot-spot regions. Referrals were received from clinical geneticists and plastic surgeons. Regions previously tested included FGFR1 exon 7 (NM_023110.2), FGFR2 exons 8, 10, and 11 (NM_000141.4), and FGFR3 exons 7 and 10 (NM_000142.4). In patients with clinically suspected Saethre–Chotzen syndrome, exon 1 of TWIST1 (NM_000474.3) had also been Sanger sequenced, in conjunction with multiplex ligation-dependent probe amplification to detect copy-number changes.
Patient selection in the retrospective cohort was enriched for those with nonsyndromic coronal craniosynostosis (N = 100, 43%), but also included patients with clinically suspected Saethre–Chotzen syndrome (N = 24, 10%), other single-suture craniosynostosis (N = 18, 8%), multisutural craniosynostosis (N = 27, 11%), other syndromic craniosynostosis (N = 44, 19%), and nonspecific phenotypes (N = 20, 9%). Nonspecific phenotypes included craniosynostosis not otherwise specified, other cranial abnormalities (e.g. parieto-occipital skull defect), or clinical features suggestive of craniosynostosis or a syndromic diagnosis without further elaboration (e.g. brachycephaly, hypertelorism). Within the single-suture group, there were eight patients with metopic, eight with sagittal, and two with lambdoid craniosynostosis. The syndromic craniosynostosis group included patients with clinically suspected Crouzon, Apert, Pfeiffer, Muenke, Beare–Stevenson, Antley–Bixler, Baller–Gerold, Carpenter, and LADD (lacrimo-auriculo-dento-digital) syndromes.
The prospective cohort of 76 individuals consisted of 25 (33%) with nonsyndromic coronal craniosynostosis, 15 (20%) with other single suture craniosynostoses, 13 (17%) with multisutural craniosynostosis, 18 (24%) with syndromic craniosynostosis, and 5 (6%) with nonspecific phenotypes. Within the single suture group, there were 5 patients with metopic and 10 with sagittal craniosynostosis. The syndromic craniosynostosis group included patients with clinically suspected Crouzon, Pfeiffer, and Muenke syndromes, as well as cleidocranial dysplasia. TWIST1 sequencing and multiplex ligation-dependent probe amplification were also performed when Saethre–Chotzen syndrome was noted as a differential diagnosis on the request form.
Sequencing panel design
The genes included in the custom-designed panel were determined on the basis of an association with craniosynostosis in peer-reviewed literature, with a focus on clinical utility in the context of a typical craniofacial-clinic referral base (Table 1). The TWIST1 gene was not included on the panel owing to its GC-rich sequence and the relatively poor coverage of such regions using MPS.
DNA extraction and sequencing
DNA extraction methods for these samples included manual ammonium acetate extraction, the EZ1 DNA Blood 350 μl kit, and the QIAsymphony DNA Midi kit. Samples were quantified using the Qubit fluorometer (Life Technologies, Carlsbad, CA) and the Qubit dsDNA BR assay kit. Target regions were amplified using 10 ng of DNA with a custom designed Ion Torrent AmpliSeq panel (Table 1) and the Ion AmpliSeq Library kit V2. A half-volume reaction protocol recommended by the manufacturer was used. The panel generated 398 amplicons across 2 pools and 70 kb of target sequence. Amplicons were digested and barcoded using the Ion AmpliSeq Library kit V2 and Ion Xpress barcode adapters. Ten picomoles of each library was combined and clonally amplified on Ion Sphere Particles by emulsion polymerase chain reaction, using the Ion OneTouch 2 instrument (Life Technologies) and the Ion PI Template OT2 V3 kit. Template-positive Ion Sphere Particles were subsequently enriched using streptavidin-coupled Dynabeads and the Ion OneTouch ES instrument (Life Technologies). Enriched Ion Sphere Particles were loaded on an Ion PI Chip V2, and sequenced on the Ion Proton (Life Technologies) using the Ion PI Sequencing 200 V3 kit.
Data analysis and quality control
Primary and secondary data analysis was performed using the manufacturer's Torrent Suite software v4.2.1, including Coverage Analysis (v4.2.1.4) and Variant Caller (v4.2.1.0) plug-ins. The Variant Caller used a modified high-stringency germline configuration, with a quality-score threshold of 100, minimum coverage of 10 reads, and minimum allele frequency of 0.1. Because of a relative lack of vertical coverage in a known FGFR3 hot-spot region (exon 7), a “hot spot” file was used to circumvent quality thresholds and highlight all known pathogenic variants in this region, irrespective of variant-calling criteria across the panel. Several key parameters were identified for assessing run-and-sample data quality, including mean read length, total number of mapped reads, average base-coverage depth, percentage of assigned amplicon reads, and uniformity of base coverage. The upper and lower limits for these parameters were defined as ±2 standard deviations from the mean, and calculated from preliminary runs. Copy-number and structural variants were not analyzed.
Variants were annotated using the GEnome MINIng (v0.14.0) platform10 and the Ensembl Variant Effect Predictor (http://asia.ensembl.org/info/docs/tools/vep/index.html). These were then filtered in an SQL database to exclude variants with a population minor allele frequency greater than 0.1% in the Exome Aggregation Consortium database (v0.3.1), variants located more than 10 bp from a coding region, and variants confirmed as recurrent sequencing artifacts by confirmatory Sanger sequencing. Remaining variants were prioritized according to predicted functional impact, population allele frequency, Combined Annotation Dependent Depletion11 and other in silico prediction tools (including Grantham score, Align-GVGD, SIFT, and PolyPhen-2). Locus-specific and general database searches, online searches for relevant literature, and correlations with the stated phenotypes and family histories were performed. Community standards for the interpretation of sequence variants were used to determine variant classification.12 Novel missense variants were also evaluated using the HOPE protein analysis server (http://www.cmbi.ru.nl/hope/), which builds predictive protein models using the Yasara and WHAT IF Twinset.13,14,15 Sanger sequencing was used to confirm all pathogenic and likely pathogenic variants identified by MPS.
Results
Clinically significant variants identified in the retrospective cohort
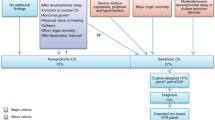
Thirty-three heterozygous non FGFR1-3 variants were identified and classified as likely pathogenic or pathogenic (Table 2). No homozygous or compound heterozygous genotypes were identified. A total of 18 variants were identified in TCF12 (55%), 6 in EFNB1 (18%), 2 in ALX4, 2 in POR, 2 in FGF10, and 1 each in EFNA4, IL11RA, and ERF. The majority of variants were identified in genes associated with autosomal dominant (73%) or X-linked phenotypes (18%). Although both POR and IL11RA are associated with autosomal recessive phenotypes, only a single pathogenic or likely pathogenic allele was identified in three patients. A second pathogenic allele was not identified in any of these individuals despite reanalysis with relaxed filtering criteria. Twenty-two variants were predicted to result in premature stop codons, 8 predicted to be missense, and 3 predicted to lead to aberrant splicing (affecting canonical splice sites ±2 bp outside coding regions). The majority of pathogenic variants were truncating or previously described missense variants. Two novel missense variants were classified as pathogenic or likely pathogenic (EFNB1 p.Leu60Pro, ALX4 p.Tyr238Asp), using criteria including absence from population databases, amino-acid conservation, location within a functional protein domain, in silico prediction scores, protein modeling software, and comparison to known variant spectra in these genes.
The total diagnostic yield in the retrospective cohort was 14% (33/233). The diagnostic yield was 16% (16/100) in individuals with non-syndromic coronal craniosynostosis, 17% (4/24) in clinically suspected Saethre–Chotzen syndrome, 6% (1/18) in other single suture craniosynostoses, 4% (1/27) in multisutural craniosynostosis, 16% (7/44) in other syndromic craniosynostoses, and 20% (4/20) in those with non-specific phenotypes.
Clinically significant variants identified in the prospective cohort
Twenty-one heterozygous pathogenic or likely pathogenic variants were detected in the prospective cohort of 76 individuals, for a diagnostic yield of 28% (Table 2). No homozygous or compound heterozygous genotypes were identified. Eleven of the variants were in FGFR1-3. The remaining 10 variants included 4 in TCF12, 2 in EFNB1, 2 in ERF, and 2 in POR (in two separate individuals). The majority of pathogenic variants were truncating or previously described missense variants. No clinically significant novel missense variants were identified. When individuals with FGFR1-3 variants were not considered, the diagnostic yield in the prospective cohort was 15%, similar to the diagnostic yield in the retrospective cohort.
Test performance characteristics
The mean number of variants identified per sample was 67 (SD ±7) after filtering by quality score alone. On average, the number of variants was then reduced to 1 (SD ±1) after filtering for variants <10 bp from a coding region with a minor allele frequency <0.1%, and excluding recurrent sequencing artifacts.
Adequate horizontal coverage was arbitrarily defined as more than 50 reads in any direction in any region. By this definition, 88% of the coding regions (±10 bp) of the genes tested had sufficient horizontal coverage. In individual genes, this value ranged from 61.5% to 100%, with a median of 94.8%. All FGFR1-3 hot-spot regions previously interrogated by Sanger sequencing met this criterion, with the exception of FGFR3 exon 7. As a result of using a “hot spot” file to prevent false-negative calls in this region, all true-positive samples were identified, together with six false-positive results. The latter represented 3% of the tested cohort. Vertical coverage metrics included a mean base-coverage depth of 6,100 reads (2 SD 1,700–10,400), uniformity of base coverage of 84.1% (2 SD 76.9–91.2), and mean percentage of assigned amplicon reads of 95.8% (2 SD 86.3–100). Mean read length across all runs was 140 bp (2 SD 130–150). For heterozygous variants deemed clinically significant and confirmed by Sanger sequencing, the proportion of MPS reads (“variant allele frequency”) for a variant ranged between 40.2% and 59.4%.
As this retrospective cohort had not been genotyped by an orthogonal method, the rate of false negatives could not be ascertained. We therefore tested an additional 24 samples with known FGFR1-3 or TWIST1 pathogenic variants, in order to compare test performance characteristics with the currently available Sanger sequencing screen. Twenty of these samples had substitution variants, and 4 samples had deletions or insertions. The sensitivity of the assay for previously identified sequence variants in FGFR1-3 was 93.8%. Two false-negative results were identified; a large 58 bp duplication (FGFR2:c.1139_1196dup58), and a complex indel (FGFR2:c.820_824delinsTT).16 The specificity of the assay was 94.6%, owing to false-positive variants in FGFR3 exon 7 resulting from use of the “hot spot” file. The positive predictive value was 83.3% and the negative predictive value was 98.1%.
Discussion
We assessed the analytical validity and clinical utility of a custom MPS gene panel for craniosynostosis in a large cohort of FGFR1-3 and TWIST1 variant-negative individuals. Pathogenic or likely pathogenic variants were identified in 14% of a retrospective cohort of 233 individuals without a prior molecular diagnosis, most commonly in genes TCF12 and EFNB1. Further unselected, prospective testing in 76 individuals showed a comparable diagnostic yield of 15% in those without variants in FGFR1-3 or TWIST1. Variants were identified most frequently in TCF12, EFNB1, and ERF.
By preemptively excluding many of the common, recurrent variants in hot-spot regions of FGFR1-3 and TWIST1, we explored whether genetic testing in additional genes would yield causative variants. Other studies have explored a similar question, but these included fewer genes and did not use MPS.4,17 There has only been one other study of a large, diverse, previously undiagnosed craniosynostosis cohort using MPS,16 which was performed by whole-exome sequencing. This approach resulted in a higher diagnostic yield of 37.5%, compared with 28% in the prospective cohort of our study; however, it is difficult to directly compare the characteristics of the two cohorts. A whole-exome or -genome approach offers the opportunity to test many more genes, and is amenable to reanalysis when additional genes are discovered later, but has the potential for discovery of incidental findings.18 A virtual panel that restricts exome data analysis to a selection of genes (such as the genes included in our panel) may be an ideal compromise between exome and panel approaches.
This study identified 22 TCF12 variants, representing 18/233 (8%) of the retrospective cohort and 4/76 (5%) of the prospective cohort. These account for 55% and 36% of all non-FGFR pathogenic variants identified in the retrospective and prospective cohorts, respectively. Our findings in the TCF12 gene are similar to those of other studies, which have identified TCF12 variants in 7% of an undifferentiated craniosynostosis cohort,17 and 6% of individuals with coronal craniosynostosis.3 We identified frameshift, nonsense, and splice-site TCF12 variants in exons 9–20 inclusive, supporting the previously noted predominance of variants in the 3′ region of the gene.3,17,19 Five individuals referred with a clinical diagnosis of Saethre–Chotzen syndrome were identified as having variants in TCF12 or EFNB1, consistent with findings of other studies.19 This confirms the difficulty of clinically identifying the causative gene for a presentation of Saethre–Chotzen syndrome without the aid of molecular testing, given its known genetic heterogeneity. It also highlights the importance of the balance between TCF12 and TWIST1 in TCF12/TWIST1 heterodimer formation and coronal suture development.3,19 These results strongly support the inclusion of TCF12 testing when pathogenic variants are not identified in FGFR1-3 and TWIST1, in individuals with nonsyndromic coronal craniosynostosis or clinically suspected Saethre–Chotzen syndrome. This is supported by other studies that assert that TCF12 testing should be performed in individuals with unilateral or bilateral coronal synostosis, and in patients with Saethre–Chotzen syndrome who are negative for TWIST1. 14,16
The relatively high number of TCF12 variants identified in the retrospective cohort may reflect the high proportion of individuals with coronal craniosynostosis or clinically suspected Saethre–Chotzen syndrome in this group (53%). This cohort is also skewed toward phenotypes associated with FGFR1-3 and TWIST1, as these were the genes being tested at the time of the original referral. This study could underestimate the diagnostic yield in individuals with noncoronal, unisutural craniosynostosis, given the lower proportion of individuals with these diagnoses in both the retrospective and the prospective cohorts.
The number of variants identified in EFNB1 was unexpected because these individuals had not been clinically diagnosed with craniofrontonasal syndrome prior to referral in the majority of cases. This suggests that EFNB1 has a wider role in undiagnosed craniosynostosis, and a broader phenotypic spectrum, than previously recognized. It is also notable that there were no clinically significant variants in 9 of the 17 non FGFR1-3 genes on the panel in the retrospective cohort, and in 12 of 17 genes in the prospective cohort. This has implications for future MPS panel design, as a more limited gene list targeted at those with the most frequent diagnoses may have the highest utility for a typical craniofacial-clinic referral base. The low diagnostic rate in both the retrospective and the prospective cohorts for unisutural noncoronal craniosynostosis is consistent with our group’s previous findings,1 and reflects that the etiology for these presentations is not completely understood.
There are some limitations to the analytical method. The deliberate omission of TWIST1 for technical reasons means that additional testing is indicated in individuals suspected to have Saethre–Chotzen syndrome; however, given that up to 28% of variants in TWIST1 are large deletions,20 additional testing for copy-number detection would nevertheless be utilized for this clinical presentation. Potentially clinically significant variants, such as noncoding variants located more than 10 bp from a coding region, could have been removed during the filtering process. Although variants with minor allele frequencies higher than 0.1% in the general population were excluded, variants leading to autosomal dominant craniosynostoses are expected to occur at a much lower population frequency and are unlikely to have been excluded. Similarly, this threshold is unlikely to influence the detection of pathogenic variants for the rare autosomal recessive phenotypes considered by this panel. Structural variants, deep intronic variants, and regulatory-region variants have not been considered, and the contribution of these mechanisms is therefore not established in this study. Alternative panel and bioinformatic analytical parameters may increase the reliability of large deletion or complex indel detection. Finally, the presence of recurrent sequencing artifacts supports the view that each laboratory should maintain its own recurrent false-positive variant data set for MPS assays.
This first study of an MPS panel for craniosynostosis has shown that testing additional genes beyond FGFR1-3 and TWIST1 can yield molecular diagnoses in up to 15% of variant-negative individuals at a moderate cost of approximately US$300. We recommend TCF12 testing in individuals with coronal synostosis or Saethre–Chotzen syndrome, either simultaneously with FGFR1-3 and TWIST1 or as a reflex test. We also recommend that an effective MPS panel for craniosynostosis should at a minimum include the genes FGFR1-3, TCF12, EFNB1, and TWIST1 (where feasible), if it is to maximize diagnostic yield while reducing cost and the likelihood of identifying variants of uncertain significance. Including ERF, ALX4, and EFNA4 may result in diagnoses in a small number of additional cases. Clinically significant variants were rarely identified in nonsyndromic, noncoronal craniosynostosis, and we do not recommend testing these genes in this population at this time. We have successfully completed clinical validation of this custom MPS craniosynostosis panel, and demonstrated that it can achieve sufficient sensitivity, specificity, and increase in diagnostic yield to have clinical utility in craniofacial clinics.
References
Roscioli T, Elakis G, Cox TC et al. Genotype and clinical care correlations in craniosynostosis: Findings from a cohort of 630 Australian and New Zealand patients. Am J Med Genet Part C Semin Med Genet 2013;163:259–270.
Johnson D, Wilkie AO. Craniosynostosis. Eur J Hum Genet 2011;19:369–376.
Sharma VP, Fenwick AL, Brockop MS et al. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat Genet 2013;45:308–313.
Wilkie AO, Byren JC, Hurst JA et al. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics 2010;126:e391–400.
Ursitti F, Fadda T, Papetti L et al. Evaluation and management of nonsyndromic craniosynostosis. Acta Paediatr 2011;100:1185–1194.
Bessenyei B, Nagy A, Szakszon K et al. Clinical and genetic characteristics of craniosynostosis in Hungary. Am J Med Genet A 2015;167A:2985–2991.
Twigg SRF, Vorgia E, McGowan SJ et al. Reduced dosage of ERF causes complex craniosynostosis in humans and mice, and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet 2013;45:304–307.
Vissers LE, Cox TC, Maga AM et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet 2011;7:e1002278.
Merrill AE, Bochukova EG, Brugger SM et al. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet 2006;15:1319–1328.
Paila U, Chapman BA, Kirchner R, Quinlan AR. GEMINI: integrative exploration of genetic variation and genome annotations. PLoS Comput Biol 2013;9:e1003153.
Kircher M, Witten DM, Jain P et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310–315.
Richards S, Aziz N, Bale S et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424.
Venselaar H, Te Beek TA, Kuipers RK et al. Protein structure analysis of mutations causing heritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548.
Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402.
Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph 1990;8:52–56.
Miller KA, Twigg SR, McGowan SJ et al. Diagnostic value of exome and whole genome sequencing in craniosynostosis. J Med Genet 2017;54:260–268.
Paumard-Hernandez B, Berges-Soria J, Barroso E et al. Expanding the mutation spectrum in 182 Spanish probands with craniosynostosis: identification and characterisation of novel TCF12 variants. Eur J Hum Genet 2015;23:907–914.
Green RC, Berg JS, Grody WW et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574.
di Rocco F, Baujat G, Arnaud E et al. Clinical spectrum and outcomes in families with coronal synostosis and TCF12 mutations. Eur J Hum Genet 2014;22:1413–1416.
Gripp KW, Kasparcova V, McDonald-McGinn DM et al. A diagnostic approach to identifying submicroscopic 7p21 deletions in Saethre-Chotzen syndrome: Fluorescence in situ hybridization and dosage-sensitive Southern blot analysis. Genet Med. 2001;3:102–108.
Acknowledgments
We acknowledge funding provided by the University of New South Wales for laboratory infrastructure, the National Health and Medical Research Council for an overseas fellowship (T.R.), Luke Hesson for critically reviewing the manuscript, and the families and clinicians for their involvement.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Lee, E., Le, T., Zhu, Y. et al. A craniosynostosis massively parallel sequencing panel study in 309 Australian and New Zealand patients: findings and recommendations. Genet Med 20, 1061–1068 (2018). https://doi.org/10.1038/gim.2017.214
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.214
Keywords
This article is cited by
-
Clinical interest of molecular study in cases of isolated midline craniosynostosis
European Journal of Human Genetics (2023)
-
The current understanding of germline predisposition in non-syndromic sagittal craniosynostosis: a systematic review
Child's Nervous System (2023)
-
Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders
Nature Communications (2020)
-
Nonsyndromic craniosynostosis: novel coding variants
Pediatric Research (2019)