Abstract
Hypoxia-inducible factor-1α (HIF-1α) is a key transcriptional mediator that coordinates the expression of various genes involved in tumorigenesis in response to changes in oxygen tension. The stability of HIF-1α protein is determined by oxygen-dependent prolyl hydroxylation, which is required for binding of the von Hippel-Lindau protein (VHL), the recognition component of an E3 ubiquitin ligase that targets HIF-1α for ubiquitination and degradation. Here, we demonstrate that PLD2 protein itself interacts with HIF-1α, prolyl hydroxylase (PHD) and VHL to promote degradation of HIF-1α via the proteasomal pathway independent of lipase activity. PLD2 increases PHD2-mediated hydroxylation of HIF-1α by increasing the interaction of HIF-1α with PHD2. Moreover, PLD2 promotes VHL-dependent HIF-1α degradation by accelerating the association between VHL and HIF-1α. The interaction of the pleckstrin homology domain of PLD2 with HIF-1α also promoted degradation of HIF-1α and decreased expression of its target genes. These results indicate that PLD2 negatively regulates the stability of HIF-1α through the dynamic assembly of HIF-1α, PHD2 and VHL.
Similar content being viewed by others
Introduction
Phospholipase D (PLD), which provides survival signaling in various cancer cells, has been reported to be elevated in many human cancers.1, 2 The overexpression of PLD promotes tumor growth and metastasis, whereas targeting of PLD suppresses tumorigenesis.3, 4, 5, 6, 7 PLD catalyzes the hydrolysis of phospholipids to generate phosphatidic acid, which consequently activates a diverse signaling cascade.2 There are two mammalian isoforms, PLD1 and PLD2, which have several conserved regions including the phox and pleckstrin homology (PH) domains as well as two catalytic regions (histidine-lysine-aspartic acid (HKD) motifs).8, 9 Although PLD activity is involved in a variety of physiological functions including proliferation, survival, invasion and differentiation, the PLD protein itself has pivotal roles in the regulation of its biological functions through interaction with signaling biomolecules independent of lipase activity. 10, 11, 12, 13
Hypoxia-inducible factor 1 (HIF-1) is a master transcriptional regulator that mediates changes in gene expression that are essential for adaptive responses during hypoxia.14, 15 Hypoxia is a typical feature of solid tumors. HIF1-α is a major survival factor for tumor cells growing in low oxygen environments. Under normoxic conditions, HIF-1α is degraded via the von Hippel-Lindau protein (VHL)-mediated ubiquitin-proteasome pathway.16 The association of VHL and HIF-1α is triggered by hydroxylation of the two proline residues (Pro402 and Pro564) within the oxygen-dependent degradation domain of HIF-1α through prolyl hydroxylase (PHD).17 Under hypoxic conditions, the activity of the PHD is inhibited, which results in the stabilization of HIF-1α. HIF-1α dimerizes with HIF-1β and binds to hypoxia-response elements on promoters of its target genes, which stimulates transcription of genes that regulate angiogenesis and other factors important for responding to hypoxic conditions such as vascular endothelial growth factor (VEGF) and glycolytic enzymes.18, 19 The functions of these genes include cell growth promotion, cell transformation, angiogenesis, apoptosis and energy metabolism.20 PLD was recently reported to regulate HIF-1α, and its activity is required for the upregulation of HIF-1α expression at the translational level.21, 22, 23 PLD1 activity is also involved in IL-1β-induced expression of angiogenic factors via the binding of HIF-1α to the promoters of its target genes.24 Moreover, HIF-1α upregulates PLD2 expression in response to hypoxic stress.25 Thus, PLD and HIF-1α might be involved in tumor progression via positive feedback regulation. Interestingly, we found that PLD1 protein itself mediates efficient degradation of HIF-1α by coordinating the dynamic assembly of PHD-HIF-1α-VHL.23 In the present study, we demonstrate that PLD2 also acts as a bridge molecule, interacting with HIF-1α, PHD and VHL, thereby assembling a multiprotein complex that destabilizes HIF-1α independent of PLD activity.
Materials and Methods
Cell culture and transfection
HEK293 and DLD1 cells were cultured in Dulbecco’s modified Eagle’s medium with 10% heat-inactivated fetal bovine serum with 1% antibiotic-antimycotic (Gibco BRL, Grand Island, NY, USA) at 37 °C in a 5% CO2/95% air incubator. Hypoxia was induced by exposing cells to 1% O2/5% CO2 balanced with N2 in a hypoxic chamber (Forma, Waltham, MA, USA). Cells were transfected using Lipofectamine Plus or Lipofectamine 2000 reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions.
Immunoblot and immunoprecipitation
Cells were lysed using passive lysis buffer (Promega, Madison, WI, USA) followed by gentle sonication. Cell lysates were incubated with suitable antibody in the presence of protein A-Sepharose beads (Amersham, Piscataway, NJ, USA). Immunoprecipitates and cell lysates were boiled with sodium dodecyl sulfate (SDS) sample buffer and then separated by SDS-polyacrylamide gel electrophoresis. The antibodies used in immunoblot and immunoprecipitation were: HIF-1α, α-tubulin, GFP, hemagglutinin (HA), myc epitope, glutathione-S-transferase (GST), VEGF, glucose transporter 3 (GLUT3) and hexokinase II (Santa Cruz Biotechnology, Santa Cruz, CA, USA); and HIF-1α (BD Bioscience, Rockville, MD, USA). A polyclonal anti-PLD antibody that recognizes both PLD1 and PLD2 was generated as previously described.26
GST pull-down assay
GST-fusion proteins were expressed using Escherichia coli BL21 cells. BL21 cells harboring each of the GST-fusion vectors were grown with 0.5 mM IPTG for 6 h at 25 °C with shaking at 200 rpm. Bacterial lysates were then applied to glutathione–sepharose 4B beads (Amersham) and washed five times with phosphate-buffered saline (containing 1% Triton X-100). In vitro-translated proteins were generated using the TNT quick coupled transcription/translation system (Promega). Glutathione–sepharose bead-conjugated GST-fusion proteins were mixed with cell lysates or in vitro-translated proteins at 4 °C for 1 h, then washed five times with phosphate-buffered saline (containing 1% Triton X-100). Proteins were eluted in SDS sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis.
Luciferase assay
The HEK293 cells were seeded in 24-well plates and then transiently transfected with pGL2-hypoxia-response elements-Luc and pRL-TK (internal control). The activities of Firefly and Renilla luciferase in the cell extracts were subsequently measured using a Dual-Luciferase Assay kit (Promega) according to the manufacturer’s instructions.
Gelatin zymography
DLD1 cells were transfected with GFP-PLD2-PH for 48 h then refreshed with serum-free Dulbecco’s modified Eagle’s medium. The activity of electrophoretically separated gelatinolytic enzymes in the conditioned media was then analyzed. In brief, equal amounts of conditioned media were mixed with 2 × zymogram buffer and then electrophoresed on SDS-polyacrylamide gel electrophoresis gels containing 1 mg ml−1 gelatin. Following electrophoresis, the gels were washed with 2.5% Triton X-100 and incubated with developing buffer, then stained with 2% Coomassie brilliant blue solution. Zones of gelatinolytic activity were detected as clear bands on a dark background. Inverted images are presented.
Statistics
The results are expressed as the mean±s.d. of the number of determinations indicated. Statistical significance of differences was determined by analysis of variance. A P<0.05 or 0.01 was considered to indicate significance.
Results
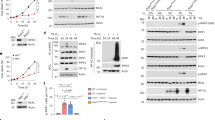
PLD2 protein promotes degradation of HIF-1α protein via the proteasomal pathway
Because PLD1 was reported to promote degradation of HIF-1α protein,23 we investigated whether PLD2 is involved in the destabilization of HIF-1α. HIF-1α was detected at very low levels under normoxia; thus, HEK293 cells were switched from hypoxia to normoxia (reoxygenation) to enable easy detection of the change in HIF-1α protein. The reoxygenated cells were treated with cycloheximide (CHX) to block de novo protein synthesis. PLD2 increased degradation of endogenous HIF-1α protein from a half-life of ~23–10 min, compared with that of the empty vector (Figure 1a). Ectopic expression of PLD2 shows an increased basal level of HIF-1α relative to that of the control (Figure 1a). Catalytically inactive mutant (mt) of PLD2 also increased destabilization of HIF-1α in the presence of CHX under reoxygenation conditions (Figure 1b), suggesting that PLD2 reduces the stability of HIF-1α independent of lipase activity. As a control, CHX blocked new protein synthesis. MtPLD2 further promoted degradation of HIF-1α with treatment of CHX relative to either case (Figure 1b). We next tried to investigate whether PLD2-mediated degradation of HIF-1α occurred via the ubiquitin-proteasome pathway, which is the main degradation pathway for HIF-1α. The proteasome inhibitor MG132 suppressed PLD2-mediated degradation of endogenous HIF-1α (Figure 1c). Moreover, both wild-type (wt) and mt PLD2 increased ubiquitination of endogenous and exogenous HIF-1α in the presence of MG132 (Figure 1d). Collectively, these results show that PLD2 promotes HIF-1α degradation via the proteosomal pathway, independent of its enzymatic activity.
PLD2 protein promotes degradation of HIF-1α protein via the proteasomal pathway. (a) HEK293 cells were transfected with vector or PLD2 and incubated under hypoxia (1% O2) for 4 h and then subjected to reoxygenation (21% O2) and treatment with CHX (100 μg ml−1) for the indicated time. The level of endogenous HIF-1α was analyzed by immunoblotting. The band intensity was quantified, and the levels of HIF-1α were normalized against α-tubulin. (b) HEK293 cells were transfected with mtPLD2 and incubated under hypoxia for 4 h, after which they were reoxygenated by treatment with CHX for 30 min. The level of endogenous HIF-1α was analyzed by immunoblotting, after which the band intensity was quantified. (c) HEK293 cells were transfected with PLD2 and incubated under hypoxia for 4 h then reoxygenated by treatment with CHX and/or MG132 for 30 min. The lysates were analyzed by immunoblotting, and the band intensity was quantified. (d) HEK293 cells were co-transfected with the indicated constructs in the presence of MG132, after which immunoprecipitation was performed to measure the ubiquitination of HIF-1α. The data are representative of three independent experiments.
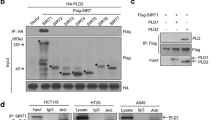
PLD2 directly interacts with HIF-1α
The stability of HIF-1α is regulated by physical interactions with various proteins.27 We recently reported that PLD1 protein itself reduces the stability of HIF-1α via its association with HIF-1α.23 Thus, we examined whether PLD2-mediated destabilization of HIF-1α occurs through their interaction. A coimmunoprecipitation experiment showed that PLD2 associates with HIF-1α (Figure 2a). To determine whether the interaction between PLD2 and HIF-1α is direct, we conducted an in vitro binding assay using in vitro translated HA-HIF-1α and PLD2. PLD2 immunoprecipitates, but not immunoglobulin G, contained HIF-1α, indicating a direct interaction between HIF-1α and PLD2 (Figure 2b). We also performed a GST pull-down assay using GST-PLD2 fragment fusion proteins to identify domain(s) of PLD2 that interact with HIF-1α. The in vitro GST pull-down assay revealed that HIF-1α binds to GST-PLD2-PH (amino acids 201–310) and GST-PLD2-F3 (amino acids 476– 612) but not GST alone (Figure 2c). These data indicate that PLD2 associates directly with HIF-1α.
PLD2 directly interacts with HIF-1α. (a) HEK293 cells were transfected with the indicated constructs, and coimmunoprecipitation was performed to examine the interaction of HIF-1α with PLD2 in the presence of MG132 (2.5 mM) for 6 h. (b) Immunoprecipitation assay of in vitro translated-HA-HIF-1α and PLD2. (c) GST pull-down assay of binding domain mapping of PLD2. A schematic representation of the PLD1 domain is shown. The data are representative of three independent experiments.
PLD2 promotes PHD2-mediated hydroxylation of HIF-1α by increasing the interaction of HIF-1α with PHD2
PHD-mediated prolyl hydroxylation of HIF-1α triggers proteasomal degradation of HIF-1α, leading to VHL-dependent ubiquitination and proteasomal degradation. Under normoxic conditions, HIF-1α is hydroxylated at proline 402 and 564. As PHD2 has a predominant role in the hydroxylation of HIF-1α, but PHD1 and PHD3 are more active on HIF-2α,28, 29 PHD2 was used for subsequent experiments. In the presence of CHX, PHD2 or PLD2 stimulated degradation of endogenous HIF-1α under reoxygenation conditions, whereas coexpression of PLD2 with PHD2 further enhanced the degradation of HIF-1α (Figure 3a). We also examined whether PLD2-mediated HIF-1α degradation required the hydroxylation of HIF-1α by PHD2. HEK293 cells were transfected with PLD2 and/or PHD2 and then treated with MG132. PHD2 or PLD2 promoted prolyl hydroxylation of endogenous HIF-1α, which was potentiated by coexpression of PLD2 with PHD2 (Figure 3b). PLD2 might increase the rate of prolyl hydroxylation by enhancing the interaction of HIF-1α with PHD2 or interacting with both HIF-1α and PHD2. This hypothesis was supported by the finding that PLD2 increased the interaction of PHD2 with HIF-1α (Figure 3c) and also interacted with PHD2 (Figure 3d). To verify that the interaction between PLD2 and PHD2 is direct, we conducted in vitro binding assays using in vitro translated PLD2 and HA-PHD2. PLD2 coimmunoprecipitated with PHD2, indicating a direct interaction between PHD2 and PLD2 (Figure 3e). Furthermore, PHD2 immunoprecipitated HIF-1α and PLD2, whereas PLD2 in the eluate of the PHD immune complex also immunoprecipitated HIF-1α (Figure 3f). Taken together, these findings support the existence of a HIF-1α-PLD2-PHD2 triple complex. These results suggest that PLD2 promotes PHD2-mediated hydroxylation of HIF-1α by increasing the interaction of HIF-1α with PHD2.
PLD2 promotes PHD2-mediated hydroxylation of HIF-1α by increasing the interaction of HIF-1α with PHD2. (a) The effect of PLD2 and/or PHD2 on the stability of HIF-1α. HEK293 cells were co-transfected with the indicated constructs and incubated under hypoxia (1% O2) for 4 h then reoxygenated (21% O2) for 30 min while being treated with CHX (100 μg ml−1). The level of endogenous HIF-1α was analyzed by immunoblotting. The band intensity was quantified, and the levels of HIF-1α were normalized against α-tubulin. (b) Immunoblot assay of hyroxylated-HIF-1α in HEK293 cells co-transfected with the indicated constructs and incubated with MG132 for 6 h. The levels of hydroxylated HIF-1α were normalized against the total HIF-1α. (c) Effect of PLD2 on the interaction of HIF-1α with PHD2. (d) Immunoprecipitation assay for the binding of PLD2 with PHD2. (e) Immunoprecipitation assay of in vitro translated-HA-PHD2 and PLD2. (f) PLD2 forms triple complexes with PHD2 and HIF-1α. The cells were co-transfected with the indicated constructs. Coimmunoprecipitation assay of lysates prepared from HEK293 cells in the presence of MG132. Lysates were immunoprecipitated with anti-GFP antibody and immunoblotted with the indicated antibody. The proteins released from primary immunoprecipitates were reimmunoprecipitated with PLD antibody and analyzed by immunoblot with anti-HIF-1α. The data are representative of three independent experiments.
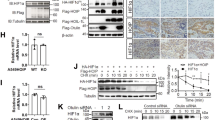
PLD2 promotes VHL-mediated HIF-1α degradation through interaction of VHL with HIF-1α
Because VHL is responsible for the degradation of HIF-1α via its interaction with HIF-1α, we investigated whether PLD2 had any effect on the VHL-mediated degradation of HIF-1α. Under reoxygenation conditions in the presence of CHX, PLD2 promoted degradation of endogenous HIF-1α, which was further potentiated by VHL (Figure 4a), indicating that PLD2 cooperates with VHL to enhance VHL-mediated HIF-1α degradation. Furthermore, the level of HIF-1α was not affected by PLD2 in VHL null renal carcimoma cells (UMRC) (Figure 4b), indicating that VHL is required for PLD2-mediated HIF degradation. We further explored the possibility that PLD2 promotes the binding of VHL with HIF-1α. PLD2 enhanced the interaction of VHL with HIF-1α (Figure 4c). To further investigate how PLD2 cooperates with VHL to destabilize HIF-1α protein, we examined whether PLD2 interacts with VHL. PLD2 was coimmunoprecipitated with VHL independent of the lipase activity (Figure 4d). GST pull-down assays using GST-VHL and in vitro translated PLD2 revealed that PLD2 interacted directly with VHL (Figure 4e). We next analyzed the binding domain required for their interaction. The β-domain of VHL (residue 1 to 154) was associated with PLD2 (Figure 4f), whereas GST-PLD2-PH (residues 201–310) bound to VHL (Figure 4g). These results suggest that PLD2 promotes VHL-dependent HIF-1α degradation by accelerating the association between VHL and HIF-1α.
PLD2 promotes VHL-mediated HIF-1α degradation through the interaction of HIF-1α with VHL. (a) Effect of PLD2 on VHL-mediated HIF-1α degradation. HEK293 cells were transfected with the indicated constructs and incubated under reoxygenated conditions with treatment of CHX for 30 min. Lysates were analyzed by immunoblot, and the band intensity was quantified. The levels of endogenous HIF-1α were normalized against α-tubulin. (b) Immunoblot assay of endogenous HIF-1α in VHL null renal carcimoma cells transfected with PLD2 or empty vector in the presence of CHX. (c) Immunoprecipitation assay of the effect of PLD2 on the interaction of VHL with HIF-1α. (d) Immunoprecipitation assay for the interaction of VHL with wt or mtPLD2. (e) GST pull-down assays of the binding of in vitro translated PLD2 with GST-VHL. (f) Domain mapping of VHL interacting with PLD2. HEK293 cells were co-transfected with PLD2 and HA-VHL fragments. Lysates were immunoprecipitated with anti-HA and immunoblotted with antibody to PLD2 antibody. (g) GST pull-down assay for domain mapping of PLD2 associated with VHL. The data are representative of three independent experiments.
PLD2-PH suppresses transactivation of HIF-1α via degradation of HIF-1α
We further examined whether the PLD2-PH domain itself, which binds to HIF-1α and VHL, could act as a regulator of stability of HIF-1α like intact PLD2. As expected, PLD2-PH promoted degradation HIF-1α under reoxygenation conditions, compared with that of empty vector (Figure 5a). Moreover, ectopic expression of PLD2-PH in DLD1 colorectal cancer cells reduced the expression of HIF-1α and its target genes, including VEGF, glucose transporter 3 and hexokinase II, under hypoxic conditions (Figure 5b). In addition, PLD2-PH significantly suppressed transactivation of HIF-1α under both normoxic and hypoxic conditions as analyzed by a hypoxia-response elements-luciferase assay (Figure 5c). Furthermore, a zymography assay revealed that PLD2-PH abolished the activity of matrix metalloproteinase 2, which is involved in the invasion capability of cancer cells (Figure 5d). Taken together, these results suggest that PLD2-PH reduces transactivation of HIF-1α and expression of its target genes via the degradation of HIF-1α.
The PH domain of PLD2 suppresses transactivation of HIF-1α via degradation of HIF-1α. (a) Effect of the PH domain of PLD2 on the stability of HIF-1α. HEK293 cells were transfected with PLD2-PH and incubated under hypoxia for 4 h then reoxygenated for 30 min with CHX for the indicated times, after which the band intensity was quantified. (b) Effect of PLD2-PH on the expression of HIF-1α target genes under hypoxia. (c) Effect of PLD2-PH on the HIF-1α transactivation of HRE under normoxia and hypoxia. *P<0.05 versus vector. (d) Gelatin zymography assay of MMP-2 under hypoxia. DLD1 cells were transfected with GFP-PLD2-PH and incubated under hypoxia. The conditioned media were concentrated and subjected to zymography.
Discussion
In the present study, we demonstrated that PLD2 has a pivotal role in the regulation of the stability of HIF-1α protein. PLD2 protein promotes the degradation of HIF-1α by interacting with HIF-1α, PHD and VHL, independent of its lipase activity. It has been reported that PLD activity is required for the upregulation of HIF-1α through increased translation21, 22, 23 but that PLD1 protein itself negatively regulates the stability of HIF-1α,23 suggesting that both PLD1 and PLD2 isozymes have dual roles in the regulation of HIF-1α. Interestingly, it has been reported that HIF-1α upregulates PLD2 at the transcriptional level under hypoxic conditions, although it is unknown how HIF-1α induces PLD2 expression.25 Thus, PLD and HIF-1α might affect their biological functions via reciprocal regulation. Because PLD mediates mitogenic and oncogenic signaling1 and HIF-1α is a central initiator of angiogenic activity in tumors, it was suggested that PLD might utilize the HIF-1α-VEGF pathway as a tumorigenic effector.22
Although PLD-mediated physiological functions are primarily induced by the product of its activity, that is, phosphatidic acid, there is a growing body of evidence that PLD protein is involved in the regulation of biological functions through binding signaling molecules independent of lipase activity. The phox domain of PLD2 acts as both a GTPase activating protein for dynamin11 and a potent guanine nucleotide exchange factor for many small GTPases regardless of its activity.12 The PH domains of PLD1 and PLD2 have been implicated as modulators of the membrane recycling machinery that result in regulated growth factor receptor endocytosis and are linked to protein–protein interactions in cellular signaling.30, 31 PLD2 promotes recruitment of PHD2 to HIF-1α and enhances prolyl hydroxylation of HIF-1α. PLD2 might provide a binding site to bring HIF-1α and PHD in proximity to each other, leading to effective hydroxylation of HIF-1α. VHL is known to recognize hydroxylated HIF-1α, leading to polyubiquitination and proteasomal degradation. Moreover, PLD2 is associated with VHL and promotes the interaction between VHL and HIF-1α, followed by degradation of HIF-1α. The molecular scaffold function of PLD2 is comparable to that of PLD1.23 Although PLD1 activity is required to enhance translation of HIF-α, PLD1 protein itself acts as a molecular scaffold, interacting directly with HIF-1α, PHD and VHL, and thereby dynamically assembling a multiprotein complex that mediates efficient degradation of HIF-1α.23 Thus, PLD1 and PLD2 isozymes might coordinate the function of HIF-1α as a hypoxic regulator in which hydroxylation and ubiquitylation are intimately associated enzymatic activities in a complex. PLD2 also plays a novel role in the canonical pathway of HIF-1α degradation.
When PLD1 or PLD2 is overexpressed in cancer cells, the net level of HIF-1α is increased despite the degradation of HIF-1α protein by PLD protein itself. Thus, it seems that PLD activity-mediated HIF-1α translation may predominantly contribute to the increased level of HIF-1α, despite the negative regulation of HIF-1α induced by PLD protein.
Under hypoxic conditions, PLD2-PH itself promotes degradation of HIF-1α and reduces expression of its target genes such as VEGF, which are comparable to the effects of PLD1-PH. Thus, the PH domain of PLD isozymes might be useful in the development of therapeutics targeting HIF-1α in cancers with tumor hypoxia.
References
Kang DW, Choi KY, Min DS . Functional regulation of phospholipase D expression in cancer and inflammation. J Biol Chem 2014; 289: 22575–22582.
Bruntz RC, Lindsley CW, Brown HA . Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacol Rev 2014; 66: 1033–1079.
Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA et al. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Sci Signal 2012; 5: ra79.
Jang YH, Choi KY, Min DS . Phospholipase D-mediated autophagic regulation is a potential target for cancer therapy. Cell Death Differ 2014; 21: 533–546.
Kang DW, Choi CY, Cho YH, Tian H, Di Paolo G, Choi KY et al. Targeting phospholipase D1 attenuates intestinal tumorigenesis by controlling β-catenin signaling in cancer-initiating cells. J Exp Med 2015; 212: 1219–1237.
Henkels KM, Boivin GP, Dudley ES, Berberich SJ, Gomez-Cambronero J . Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene 2013; 32: 5551–5562.
Gomez-Cambronero J . Phospholipase D in cell signaling: from a myriad of cell functions to cancer growth and metastasis. J Biol Chem 2014; 286: 22557–22566.
Hammond SM, Altshuller YM, Sung TC, Rudge SA, Rose K, Engebrecht J, Morris AJ et al. Human ADP-ribosylation factor-activated phosphatidylcholine-specific phospholipase D defines a new and highly conserved gene family. J Biol Chem 1995; 270: 29640–29643.
Colley WC, Altshuller YM, Sue-Ling CK, Copeland NG, Gilbert DJ, Jenkins NA et al. Cloning and expression analysis of murine phospholipase D1. Biochem J 1997; 326: 745–753.
Jang JH, Lee CS, Hwang D, Ryu SH . Understanding of the roles of phospholipase D and phosphatidic acid through their binding partners. Prog Lipid Res 2012; 51: 71–81.
Lee CS, Kim IS, Park JB, Lee MN, Lee HY, Suh PG et al. The phox homology domain of phospholipase D activates dynamin GTPase activity and accelerates EGFR endocytosis. Nat Cell Biol 2006; 8: 477–484.
Gomez-Cambronero J . Biochemical and cellular implications of a dual lipase-GEF function of phospholipase D2. J Leukoc Biol 2012; 92: 461–467.
Mahankali M, Henkels KM, Alter G, Gomez-Cambronero J . Identification of the catalytic site of phospholipase D2 (PLD2) newly described guanine nucleotide exchange factor activity. J Biol Chem 2012; 287: 41417–41431.
Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 1998; 12: 149–162.
Yu AY, Shimoda LA, Iyer NV, Huso DL, Sun X, McWilliams R et al. Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1alpha. J Clin Invest 1999; 103: 691–696.
Salceda S, Caro J . Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem 1997; 272: 22642–22647.
Yee Koh M, Spivak-Kroizman TR, Powis G . HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci 2008; 33: 526–534.
Shaw RJ . Glucose metabolism and cancer. Curr Opin Cell Biol 2006; 18: 598–608.
Semenza GL . Intratumoral hypoxia, radiation resistance, and HIF-1. Cancer Cell 2004; 5: 405–406.
Harris AL . Hypoxia-a key regulatory factor in tumor growth. Nat Rev Cancer 2002; 2: 405–406.
Toschi A, Edelstein J, Rockwell P, Ohh M, Foster DA . HIF alpha expression in VHL-deficient renal cancer cells is dependent on phospholipase D. Oncogene 2008; 27: 2746–2753.
Han S, Huh J, Kim W, Jeong S, Min DS, Jung Y . Phospholipase D activates HIF-1-VEGF pathway via phosphatidic acid. Exp Mol Med 2014; 46: 126.
Park MH, Choi KY, Jung Y, Min DS . Phospholipase D1 protein coordinates dynamic assembly of HIF-1α-PHD-VHL to regulate HIF-1α stability. Oncotarget 2014; 5: 11857.
Kang DW, Park MK, Oh HJ, Lee DG, Park SH, Choi KY et al. Phospholipase D1 has a pivotal role in interleukin-1β-driven chronic autoimmune arthritis through regulation of NF-κB, hypoxia-inducible factor 1α, and FoxO3a. Mol Cell Biol 2013; 33: 2760–2772.
Liu M, Du K, Fu Z, Zhang S, Wu X . Hypoxia-inducible for 1-alpha up-regulates the expression of phospholipase D2 in colon cancer cells under hypoxic conditions. Med Oncol 2015; 32: 394–403.
Min DS, Ahn BH, Rhie DJ, Yoon SH, Hahn SJ, Kim MS et al. Expression and regulation of phospholipase D during neuronal differentiation of PC12 cells. Neuropharmacology 2001; 41: 384–391.
Majmundar AJ, Wong WJ, Simon MC . Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 2010; 40: 294–309.
Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J . HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J 2003; 22: 4082–4090.
Webb JD, Coleman ML, Pugh CW . Hypoxia, hypoxia-inducible factors (HIF), HIF hydroxylases and oxygen sensing. Cell Mol Life Sci 2009; 66: 3539–3554.
Gomez-Cambronero J . Phosphatidic acid, phospholipase D and tumorigenesis. Adv Biol Regul 2014; 54: 197–206.
Lemmon MA, Ferguson KM . Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 2000; 350: 1–18.
Acknowledgements
This study was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (NRF-2015R1A2A1A05001884) and a Translational Research Center for Protein Function Control Grant (NSF 2009-0092960).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Park, M., Bae, S., Choi, KY. et al. Phospholipase D2 promotes degradation of hypoxia-inducible factor-1α independent of lipase activity. Exp Mol Med 47, e196 (2015). https://doi.org/10.1038/emm.2015.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2015.87
This article is cited by
-
Phospholipase D2 is a positive regulator of sirtuin 1 and modulates p53-mediated apoptosis via sirtuin 1
Experimental & Molecular Medicine (2021)