Abstract
MicroRNAs (miRNAs) regulate the proliferation and metastasis of cancer cells. Here, we showed that miR-152 was downregulated in non-small-cell lung cancer (NSCLC) tissues and cell lines. Overexpression of miR-152 suppressed cell proliferation and colony formation and also limited migration and invasion. Fibroblast growth factor 2 (FGF2) was confirmed as a direct target of miR-152. FGF2 knockdown suppressed cell proliferation, colony formation, migration and invasion, whereas FGF2 overexpression partially reversed the suppressive effect of miR-152. Furthermore, the presence of miR-152 was inversely correlated with FGF2 in NSCLC tissues. Overall, this study demonstrated that miR-152 suppressed the proliferation and invasion of NSCLC cells by downregulating FGF2. These findings provide novel insights with potential therapeutic applications for the treatment of NSCLC.
Similar content being viewed by others
Introduction
Lung cancer is the most common cause of cancer-related death worldwide. Its incidence is rapidly increasing in developing countries, with non-small-cell lung cancer (NSCLC) accounting for >80% of all lung cancer cases.1 The prognosis for NSCLC remains poor despite recent advances in the diagnosis of and chemotherapies used for this cancer, and the 5-year overall survival rate of NSCLC is a dismal 11%.2 Thus, the elucidation of the molecular mechanisms that control NSCLC tumor metastasis is urgently needed.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that negatively regulate the expression of their target genes by binding to the 3′-untranslated regions (3′-UTRs) of target mRNAs that leads to mRNA degradation or translational suppression.3, 4 The miRNAs regulate the expression of multiple target genes involved in various biological processes, including cell proliferation, differentiation, migration and apoptosis.5, 6 Recently, mounting evidence has indicated that aberrant changes in miRNA expression correlates with a wide range of cancers and that miRNAs act as oncogenes and tumor suppressors.7, 8 In NSCLC, multiple miRNAs, such as miR-10b, miR-150 and miR-205, were found to promote NSCLC carcinogenesis.9, 10, 11 In contrast, miR-16, miR-140 and miR-223 have been identified as tumor suppressors.7, 12, 13 In several cancers, including NSCLC,14 miR-152 levels are decreased, whereas miR-152 functions as a tumor suppressor in cancers including prostate cancer, glioma and endometrial cancer.15, 16, 17 Recently, Su et al.18 showed that miR-152 suppresses NSCLC progression by targeting ADAM metallopeptidase domain 17 (ADAM17). However, the function of miR-152 in NSCLC remains poorly understood.
In the present study, we found that miR-152 was significantly decreased in NSCLC, whereas miR-152 overexpression significantly suppressed the proliferation and motility of NSCLC cells. Furthermore, we found that miR-152 directly targeted fibroblast growth factor 2 (FGF2) in NSCLC cells and that miR-152 was inversely correlated with FGF2 in NSCLC tissues. Together, these results suggest that miR-152 could be a therapeutic and prognostic target for the treatment of NSCLC.
Materials and methods
Patient samples and cell lines
Human NSCLCs and matched normal tissues were obtained from 30 patients in our department. Informed written consent was obtained from all patients, and this study was approved by the Medical Ethics Committee at Zhongnan Hospital. All tissue samples were stored in liquid nitrogen until RNA extraction and western blotting. Clinicopathologic information is summarized in Table 1. The NSCLC cell lines A549, SK-MES-1, H460 and H520 and the normal lung bronchus epithelial cell line 16HBE were obtained from ATCC (Manassas, VA, USA) and cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. All cells were maintained in a humidified, 5% CO2 atmosphere at 37 °C. After the NSCLC cells reached 80% confluence, they were transfected with miR-152 or control mimics using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Plasmids and luciferase activity assays
MiR-152 and control mimics/inhibitors were purchased from Ribio Bio (Guangzhou, China). FGF2 short hairpin RNA (shRNA) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). For FGF2 overexpression, the coding sequence of FGF2 was PCR-amplified and cloned into pcDNA4.0. For measuring the effect of miR-152 on the 3′-UTR of FGF2, we generated a luciferase expression construct containing part of FGF2 3′-UTR. We amplified the wild-type fragment 2132-3768 of FGF2 mRNA that contained potential miR-152-binding sites at position 3099–3121, using the following primers: 5′-GGGGTACCAGTGAGCCATAATCGTGC-3′ (forward) and 5′-CCCTCGAGAACAACTCACCTACCCAGA-3′ (reverse). The PCR fragment was inserted into the pGL3 Basic Vector using the KpnI/XhoI endonuclease restriction sites. Mutation of the FGF 3′-UTR (Mut) was performed using a mutation kit (Stratagene, La Jolla, CA, USA).
For luciferase activity assays, 50% confluent HEK293 cells were co-transfected with 100 ng of wild-type or Mut FGF2 3′-UTR and 100 nM miR-152 or control mimics using Lipofectamine 2000. Cells were also transformed with 100 ng of PGL3 control vector to monitor transfection efficiency. MiR-NC was used as a nontargeting negative control. In addition, all cells were transfected with pRL-TK (Promega, Madison, WI, USA) as a control for normalization. Cells were harvested 48 h after transfection for luciferase activity assays using the dual-luciferase reporter assay system (Promega). Relative firefly luciferase activity was obtained by normalization to Renilla luciferase activity.
RNA extraction and quantitative real-time PCR (qRT-PCR)
RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s protocol. FGF2 expression was detected with SYBR Green reagents (TAKARA, Tokyo, Japan) using the following primers: 5′-AGGAGAGCGACCCACACATCAA-3′ (forward) and 5′-AGCCAGCAGTCTTCCATCTTCC-3′ (reverse). MiRNA was extracted using an All-in-One microRNA extraction kit and detected with an All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Carlsbad, CA, USA) using SYBR Green reagents. Primers for miR-152 (Cat No. HmiRQP3058) and U6 (HmiRQP9001) were purchased from GeneCopoeia. FGF2 expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and miR-152 was normalized to U6. Expression levels were quantified using the 2−ΔΔCt method.
Cell survival assays
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8, Beyotime, Shanghai, China). Briefly, 5 × 103 cells were cultured in 96-well plates. Then, 24 h after transfection, 10 μl of CCK-8 reagent was added to each well and incubated at 37 °C for 1.5 h. Absorbance at 450 nM was detected using a microtiter plate reader.
Colony formation assay
To assess colony formation, 24 h after transfection, 500 cells were plated in 6-well plates and grown for 2 weeks; the culture medium was replaced every 4 days. Cells were fixed with methanol and stained with 0.5% crystal violet for 20 min; visible colonies were counted. Triplicate wells were measured for each group.
Flow cytometry
Cell apoptosis was assayed using flow cytometry. Briefly, treated cells were trypsinized, collected, washed and stained with Annexin V-fluorescein isothiocyanate and propidium iodide for 15 min at 4 °C. The stained cells were analyzed with a flow cytometer (FACScalibur, BD, Franklin Lakes, NJ, USA).
Migration and invasion
Migration and invasion assays were performed using Transwell chambers with a pore size of 8 μm. Cells were transfected with miR-152 or control mimics and incubated for 24 h. For migration assays, 5 × 104 transfected cells were placed in the upper chamber. Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum was added to the lower chamber as a chemoattractant. Chambers were incubated at 37 °C in 5% CO2 for 16 h, and then cells on the upper surface were removed. Cells that had migrated to the bottom surface were washed twice with cold phosphate-buffered saline, fixed in methanol and stained with 0.1% crystal violet. Stained cells were counted under a microscope. For invasion assays, the upper chamber was precoated with Matrigel. Both assays were performed in three independent experiments.
Western blotting
Cultured cells were washed three times in cold phosphate-buffered saline and lysed with RIPA buffer (pH 7.4, 50 mM Tris–HCl, 1% (v/v) Triton X-100, 1 mM leupeptin, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 1 mM Na3VO4 and 10 mM NaF). Proteins were separated by 10% SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes and incubated with primary antibodies overnight at 4 °C followed by secondary antibodies for 1 h at room temperature. Blots were visualized using an ECL detection system (Amersham, Piscataway, NJ, USA) and analyzed by Kodak Digital Science 1D software (Eastman Kodak, Rochester, NY, USA). Image J software (National institutes of Health, Bethesda, MD, USA) was used to quantify the intensity of protein bands. The relative intensity was calculated by normalization to GAPDH that was used as a loading control. Experiments were performed in triplicate.
Statistical analysis
Data are presented as the mean±s.d. Student’s t-test or analysis of variance was performed for statistical analyses using SPSS 12.0 (Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
The miR-152 levels were decreased in NSCLC tissues and cell lines
The expression of miR-152 in 30 pairs of NSCLC tissues and their matched normal tissues was measured using qRT-PCR. The results showed that miR-152 expression was significantly downregulated in NSCLC tissues compared with matched controls (Figure 1a). In addition, the expression of miR-152 in four NSCLC cell lines was determined. We found that the relative expression of miR-152 in these NSCLC cells was strikingly decreased compared with that of the normal lung bronchus epithelial cell line 16HBE (Figure 1b). These results suggest that downregulation of miR-152 may contribute to the progression of NSCLC.
MicroRNA-152 (miR-152) was decreased in non-small-cell lung cancer (NSCLC) tissues and cell lines. (a) The expression levels of miR-152 in 30 pairs of NSCLC tissues and their matched normal tissues were measured by quantitative real-time PCR (qRT-PCR). U6 was used as an internal control. (b) The expression levels of miR-152 in a normal lung epithelial cell line (16HBE) and four NSCLC cell lines (A549, SK-MES-1, H460 and H520). *P<0.05, **P<0.01 vs control.
MiR-152 suppressed proliferation of NSCLC cells
To understand the role of miR-152 in the development of NSCLC, miR-152 was transfected into A549 cells, and we then used a CCK-8 assay to determine their proliferation. We found that miR-152 overexpression significantly suppressed A549 cell proliferation compared with their corresponding controls (Figure 2a). Next, flow cytometry was used to examine the apoptosis of A549 cells, and we found that miR-152 overexpression also induced this process (Figure 2b). We also used a colony formation assay to test the possibility that alterations in gene expression could cause either cell cycle arrest or cell death and lead to a reduction in colony number. Indeed, exogenous miR-152 expression significantly inhibited A549 colony formation (Figure 2c). The expression of miR-152 after transfection with a miR-152 mimic was determined by qRT-PCR (Figure 2d). Taken together, these results suggest that miR-152 was able to suppress NSCLC cell growth and induce apoptosis.
MicroRNA-152 (miR-152) suppressed the proliferation of non-small-cell lung cancer (NSCLC) cells. (a) A549 cells were transfected with miR-152 or control mimics, and a cell viability assay (CCK-8) was performed. (b) Cell apoptosis assays. (c) Colony formation. (d) Expression of miR-152 in A549 cells transfected with miR-152 or control mimics. *P<0.05, **P<0.01 vs control.
MiR-152 suppressed NSCLC cell migration and invasion
Migration and invasion are fundamental functions underlying several cellular processes, including angiogenesis, immune response and metastasis of cancer cells. Here, Transwell chambers were used to measure the number of cells that traversed a porous membrane or moved through an extracellular matrix. These assays revealed that overexpression of miR-152 significantly decreased migration (Figure 3a) and invasion (Figure 3b) of A549 cells, suggesting that miR-152 suppressed the motility of NSCLC cells.
FGF2 was a direct target of miR-152
To identify targets of miR-152, we used TargetScan 6.2 (http://www.targetscan.org), a widely used miRNA target prediction website. FGF2 was found to be a potential target (Figure 4a). Luciferase activity assays are commonly used to validate the suppressive effects of miRNAs on their target mRNAs. Here, we found that miR-152 significantly inhibited the luciferase activity of the wild-type but not the Mut 3′-UTR of FGF2 in HEK293 cells (Figure 4b). Moreover, overexpression of miR-152 significantly suppressed levels of both FGF2 mRNA and protein, whereas the inhibition of miR-152 significantly increased these levels (Figures 4c and d). These data suggest that miR-152 might induce the degradation of FGF2 mRNA, leading to a reduction in levels of FGF2 protein and implying that FGF2 is a direct target of miR-152.
Fibroblast growth factor 2 (FGF2) was a direct target of microRNA-152 (miR-152). (a) Schematic representation of wild-type (WT) and mutated (Mut) putative miR-152-binding sites in the 3′-untranslated region (3′-UTR) of FGF2. (b) HEK293 cells were co-transfected with 100 nM WT or Mut FGF2 3'-UTR, pGL-3 control and either miR-152 or negative control mimics. Relative firefly luciferase activity was normalized to Renilla luciferase activity. (c) Expression of FGF2 mRNA was detected by quantitative real-time PCR (qRT-PCR) in A549 cells transfected with miR-152 or control mimic and in A549 cells transfected miR-152 or control inhibitor. (d) Protein levels were detected by western blot analysis in A549 cells transfected with miR-152 or control mimic and in A549 cells transfected with miR-152 or control inhibitor. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. *P<0.05, **P<0.01 vs control.
FGF2 inhibition resulted in similar effects as miR-152 overexpression
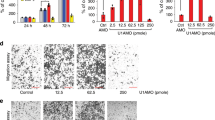
Because we found that FGF2 is a direct target of miR-152, we further investigated whether FGF2 silencing by shRNA could induce effects on NSCLC cells similar to those caused by the overexpression of miR-152. A549 cells were transfected with either an shRNA targeted against FGF2 or a nontargeting control. The inhibition of FGF2 by shRNA significantly suppressed the proliferation, migration and invasion of A549 cells (Figures 5a–c). Western blotting and qRT-PCR were used to detect the effects of FGF2 shRNA (Figures 5d and e). These data indicate that inhibition of FGF2 mimicked the effects of miR-152 overexpression.
Fibroblast growth factor 2 (FGF2) inhibition resulted in similar effects as microRNA-152 (miR-152) overexpression. (a) The viability of A549 cells transfected with FGF2 or control short hairpin RNAs (shRNAs) was detected using CCK-8. (b) In vitro migration. (c) In vitro invasion. (d) Expression of FGF2 mRNA was detected by quantitative real-time PCR (qRT-PCR) in A549 cells transfected with shRNA-NC or shRNA-FGF2. (e) FGF2 protein levels were detected by western blot analysis in A549 cells transfected with FGF2 or control shRNA. *P<0.05, **P<0.01 vs control.
FGF2 overexpression partially attenuated the tumor suppressive effect of miR-152
We further investigated whether FGF2 overexpression could attenuate the tumor suppressive effects of miR-152. Indeed, CCK-8 assays (Figure 6a) as well as in vitro migration and invasion assays (Figures 6b and c) revealed that overexpression of FGF2 significantly reversed the tumor suppressive effects of miR-152 on A549 cells. These effects were validated using another NSCLC cell line, H460. Increased levels of FGF2 mRNA after cells were transfected with an FGF2 plasmid were confirmed by qRT-PCR (Figure 6d). These data indicate that restoration of FGF2 significantly attenuated the tumor suppressive effect of miR-152.
Fibroblast growth factor 2 (FGF2) overexpression partially attenuated the tumor suppressive effect of microRNA-152 (miR-152). (a–c) A549 or H460 cells were transfected with miR-152 or control mimic (control) with or without an FGF2-overexpression plasmid. Proliferation (CCK-8 assay) (a), in vitro migration (b) and in vitro invasion (c) were evaluated as described in the Materials and methods. (d) FGF2 expression in A549 or H460 cells transfected with an FGF2-overexpression plasmid or the empty vector (control) was measured by quantitative real-time PCR (qRT-PCR). *P<0.05, **P<0.01 vs control. #P<0.05, ##P<0.01 vs miR-152 group.
FGF2 expression was inversely correlated with that of miR-152 in NSCLC tissues
To further explore the relationship between FGF2 and miR-152 in vivo, we examined the expression of FGF2 in 30 pairs of NSCLC tissues and their matched normal tissues using qRT-PCR. We found that FGF2 expression was significantly increased in NSCLC tissues relative to the matched controls (Figure 7a). Moreover, FGF2 was negatively correlated with miR-152 expression in the same NSCLC tissues (Figure 7b). These data further indicate that FGF2 was a target of miR-152 in NSCLC.
Fibroblast growth factor 2 (FGF2) levels were inversely correlated with microRNA-152 (miR-152) in non-small-cell lung cancer (NSCLC) tissues. (a) FGF2 expression levels in 30 pairs of NSCLC tissues and their matched normal tissues were measured by quantitative real-time PCR (qRT-PCR). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. (b) Correlation analysis between FGF2 mRNA levels and miR-152 expression in NSCLC tissues (Spearman’s correlation analysis, r=−0.422; P<0.05). *P<0.05 vs control.
Discussion
The miRNAs have been reported to play essential roles in carcinogenesis and tumor progression.19, 20 In addition, acting as either tumor suppressors or oncogenes, miRNAs are involved in several aspects of cancer biology including cell proliferation, apoptosis, migration and invasion.21, 22 In this study, we focused on miR-152 that belongs to the miR-148/152 family that includes miR-148a, miR-148b and miR-152. Many studies have revealed that members of the miR-148/152 family potentially act as oncogenes and tumor suppressors in different cancers.23 For example, the expression of miR-148/152 family members was found to be decreased in cholangiocarcinoma, endometrial serous adenocarcinoma, gastrointestinal cancers, hepatocellular carcinoma, oral squamous cell carcinoma, ovarian cancer, pancreatic cancer and prostate cancer, and these family members may function as tumor suppressors in these tumors.23 Azizi et al.24 found in pancreatic cancer cell lines that miR-148b and miR-152 reactivated tumor suppressor genes by inhibiting DNA methyltransferase-1 (DNMT1). Zhu et al.15 showed that overexpression of miR-152 inhibited the migratory and invasive capabilities of prostate cancer cells in vitro and that low miR-152 expression correlated with advanced pathological T stages. Moreover, Xiang et al.25 reported that overexpression of miR-152 increased the sensitivity of ovarian cancer cells to cisplatin by inhibiting proliferation and promoting apoptosis through targeting of DNMT1.
However, the role of miR-152 in NSCLC carcinogenesis remains unclear. A recent report by Su et al.18 revealed that miR-152 suppressed the proliferation, colony formation, migration and invasion of NSCLC cells by targeting ADAM17. In this study, we showed that miR-152 was markedly decreased in NSCLC tissues and cell lines. The ectopic overexpression of miR-152 effectively inhibited NSCLC cell proliferation, enhanced apoptosis and suppressed migration and invasion. These results suggest that miR-152 is a novel tumor suppressive miRNA in NSCLC. Many previous studies of miR-152 in various cancers focused on its relationship with DNA methylation.26, 27, 28 However, DNA methylation may not be the only target of miR-152, as another target molecule, ADAM17, has been recently reported. Indeed, our study suggests another miR-152 target, FGF2, in NSCLC cells.
To elucidate the molecular mechanisms involved in miR-152-induced inhibition of NSCLC growth and invasion, we used TargetScan 6.2 to predict miR-152 target genes. FGF2, which is frequently increased in many malignancies including NSCLC,29, 30, 31 was identified as a target of miR-152 in NSCLC cells. The inhibition of FGF2 expression resulted in similar effects as the overexpression of miR-152, and the restoration of FGF2 markedly attenuated the tumor suppressive effects of miR-152 on NSCLC cells. Furthermore, FGF2 levels were increased and were negatively correlated with miR-152 levels in NSCLC tissues. The miRNAs can induce mRNA degradation or translational suppression, and downregulated mRNA levels may result in decreased protein levels. Thus, the observed decrease in FGF2 protein levels induced by miR-152 might be because of the degradation of FGF2 mRNA. Together, these data suggest that miR-152 inhibits NSCLC growth and metastasis by inhibiting FGF2. In various cancers, FGF2 acts as an essential regulator of cell proliferation and metastasis.32, 33 Coleman et al.34 found that in pancreatic stellate cells, the nuclear translocation of FGF2 facilitated their invasion. Rades et al.35 reported that FGF2 was an independent prognostic factor for patients with locally advanced squamous cell carcinoma of the head and neck. FGF2 also plays essential roles in tumor angiogenesis. For example, Zhou et al.36 reported that miR-503 suppressed tumor angiogenesis and growth by targeting FGF2 and vascular endothelial growth factor A. Fons et al.37 reported that tumor vasculature could also be regulated by FGF/FGF receptor signaling-mediated angiogenesis. Here, our work further investigated the role of FGF2 in NSCLC.
In conclusion, this study demonstrated that miR-152 was significantly decreased in NSCLC tissues and cell lines. The exogenous overexpression of miR-152 inhibited tumor growth and metastasis of NSCLC cells through targeting of FGF2. Together, our data provide novel insights into therapeutic applications for treating NSCLC.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D . Global cancer statistics. CA Cancer J Clin 2011; 61: 69–90.
Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L et al. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol 2007; 8: 784–796.
Daniels MG, Bowman RV, Yang IA, Govindan R, Fong KM . An emerging place for lung cancer genomics in 2013. J Thorac Dis 2013; 5 (Suppl 5): S491–S497.
Chou J, Shahi P, Werb Z . microRNA-mediated regulation of the tumor microenvironment. Cell Cycle 2013; 12: 3262–3271.
Hu J, Fang Y, Cao Y, Qin R, Chen Q . miR-449a regulates proliferation and chemosensitivity to cisplatin by targeting cyclin D1 and BCL2 in SGC7901 cells. Dig Dis Sci 2014; 59: 336–345.
Alrfaei BM, Vemuganti R, Kuo JS . microRNA-100 targets SMRT/NCOR2, reduces proliferation, and improves survival in glioblastoma animal models. PLoS ONE 2013; 8: e80865.
Ke Y, Zhao W, Xiong J, Cao R . Downregulation of miR-16 promotes growth and motility by targeting HDGF in non-small cell lung cancer cells. FEBS Lett 2013; 587: 3153–3157.
Xin C, Zhang H, Liu Z . miR-154 suppresses colorectal cancer cell growth and motility by targeting TLR2. Mol Cell Biochem 2014; 387: 271–277.
Liu Y, Li M, Zhang G, Pang Z . MicroRNA-10b overexpression promotes non-small cell lung cancer cell proliferation and invasion. Eur J Med Res 2013; 18: 41.
Zhang N, Wei X, Xu L . miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett 2013; 587: 2346–2351.
Lei L, Huang Y, Gong W . miR-205 promotes the growth, metastasis and chemoresistance of NSCLC cells by targeting PTEN. Oncol Rep 2013; 30: 2897–2902.
Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang Y et al. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol Lett 2013; 6: 359–366.
Yuan Y, Shen Y, Xue L, Fan H . miR-140 suppresses tumor growth and metastasis of non-small cell lung cancer by targeting insulin-like growth factor 1 receptor. PLoS ONE 2013; 8: e73604.
Sanfiorenzo C, Ilie MI, Belaid A, Barlesi F, Mouroux J, Marquette CH et al. Two panels of plasma microRNAs as non-invasive biomarkers for prediction of recurrence in resectable NSCLC. PLoS ONE 2013; 8: e54596.
Zhu C, Li J, Ding Q, Cheng G, Zhou H, Tao L et al. miR-152 controls migration and invasive potential by targeting TGFalpha in prostate cancer cell lines. Prostate 2013; 73: 1082–1089.
Zheng X, Chopp M, Lu Y, Buller B, Jiang F . MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Lett 2013; 329: 146–154.
Tsuruta T, Kozaki K, Uesugi A, Furuta M, Hirasawa A, Imoto I et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res 2011; 71: 6450–6462.
Su Y, Wang Y, Zhou H, Lei L, Xu L . MicroRNA-152 targets ADAM17 to suppress NSCLC progression. FEBS Lett 2014; 588: 1983–1988.
Chira P, Vareli K, Sainis I, Papandreou C, Briasoulis E . Alterations of microRNAs in solid cancers and their prognostic value. Cancers 2010; 2: 1328–1353.
Shen Y, Tang D, Yao R, Wang M, Wang Y, Yao Y et al. microRNA expression profiles associated with survival, disease progression, and response to gefitinib in completely resected non-small-cell lung cancer with EGFR mutation. Med Oncol 2013; 30: 750.
Bi Y, Han Y, Bi H, Gao F, Wang X . miR-137 impairs the proliferative and migratory capacity of human non-small cell lung cancer cells by targeting paxillin. Hum Cell 2014; 27: 95–102.
Lei L, Huang Y, Gong W . Inhibition of miR-92b suppresses nonsmall cell lung cancer cells growth and motility by targeting RECK. Mol Cell Biochem 2014; 387: 171–176.
Chen Y, Song YX, Wang ZN . The microRNA-148/152 family: multi-faceted players. Mol Cancer 2013; 12: 43.
Azizi M, Teimoori-Toolabi L, Arzanani MK, Azadmanesh K, Fard-Esfahani P, Zeinali S . MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther 2014; 15: 419–427.
Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene 2014; 33: 378–386.
Ji W, Yang L, Yuan J, Yang L, Zhang M, Qi D et al. MicroRNA-152 targets DNA methyltransferase 1 in NiS-transformed cells via a feedback mechanism. Carcinogenesis 2013; 34: 446–453.
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol 2013; 5: 3–13.
Huang J, Wang Y, Guo Y, Sun S . Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology 2010; 52: 60–70.
Farhat FS, Tfayli A, Fakhruddin N, Mahfouz R, Otrock ZK, Alameddine RS et al. Expression, prognostic and predictive impact of VEGF and bFGF in non-small cell lung cancer. Crit Rev Oncol/Hematol 2012; 84: 149–160.
Polnaszek N, Kwabi-Addo B, Peterson LE, Ozen M, Greenberg NM, Ortega S et al. Fibroblast growth factor 2 promotes tumor progression in an autochthonous mouse model of prostate cancer. Cancer Res 2003; 63: 5754–5760.
Berger W, Setinek U, Mohr T, Kindas-Mugge I, Vetterlein M, Dekan G et al. Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int J Cancer 1999; 83: 415–423.
Lau MT, So WK, Leung PC . Fibroblast growth factor 2 induces E-cadherin down-regulation via PI3K/Akt/mTOR and MAPK/ERK signaling in ovarian cancer cells. PLoS ONE 2013; 8: e59083.
Barclay C, Li AW, Geldenhuys L, Baguma-Nibasheka M, Porter GA, Veugelers PJ et al. Basic fibroblast growth factor (FGF-2) overexpression is a risk factor for esophageal cancer recurrence and reduced survival, which is ameliorated by coexpression of the FGF-2 antisense gene. Clin Cancer Res 2005; 11: 7683–7691.
Coleman SJ, Chioni AM, Ghallab M, Anderson RK, Lemoine NR, Kocher HM et al. Nuclear translocation of FGFR1 and FGF2 in pancreatic stellate cells facilitates pancreatic cancer cell invasion. EMBO Mol Med 2014; 6: 467–481.
Rades D, Seibold ND, Gebhard MP, Noack F, Bruchhage KL, Schild SE . Fibroblast growth factor 2 is of prognostic value for patients with locally advanced squamous cell carcinoma of the head and neck. Strahlenther Onkol 2014; 190: 68–74.
Zhou B, Ma R, Si W, Li S, Xu Y, Tu X et al. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett 2013; 333: 159–169.
Fons P, Gueguen-Dorbes G, Herault JP, Geronimi F, Tuyaret J, Frederique D et al. Tumor vasculature is regulated by FGF/FGFR signaling-mediated angiogenesis and bone marrow-derived cell recruitment: this mechanism is inhibited by SSR128129E, the first allosteric antagonist of FGFRs. J Cell Physiol 2014 (e-pub ahead of print 24 April 2014; doi:10.1002/jcp.24656).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cheng, Z., Ma, R., Tan, W. et al. MiR-152 suppresses the proliferation and invasion of NSCLC cells by inhibiting FGF2. Exp Mol Med 46, e112 (2014). https://doi.org/10.1038/emm.2014.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2014.51
This article is cited by
-
MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies
Experimental & Molecular Medicine (2023)
-
Current Understanding of the Pathogenesis of Dengue Virus Infection
Current Microbiology (2021)
-
CRISPR/Cas9 – An evolving biological tool kit for cancer biology and oncology
npj Precision Oncology (2019)
-
Upregulation of miR-223 in the rat liver inhibits proliferation of hepatocytes under simulated microgravity
Experimental & Molecular Medicine (2017)
-
Long Noncoding RNA PVT1 Acts as a “Sponge” to Inhibit microRNA-152 in Gastric Cancer Cells
Digestive Diseases and Sciences (2017)