Abstract
Genetic analysis of an inbred Pakistani family PKDF280, segregating prelingual severe to profound sensorineural hearing loss, provided evidence for a DFNB locus on human chromosome 9q34.3. Co-segregation of the deafness trait with marker D9SH159 was determined by a two-point linkage analysis (LOD score 9.43 at θ=0). Two additional large families, PKDF517 and PKDF741, co-segregate recessive deafness with markers linked to the same interval. Haplotype analyses of these three families refined the interval to 3.84 Mb defined by D9S1818 (centromeric) and D9SH6 (telomeric). This interval overlaps with the previously reported DFNB33 locus whose chromosomal map position has been recently revised and assigned to a new position on chromosome 10p11.23–q21.1. The nonsyndromic deafness locus on chromosome 9q segregating in family PKDF280 was designated DFNB79. We are currently screening the 113 candidate DFNB79 genes for mutations and have excluded CACNA1B, EDF1, PTGDS, EHMT1, QSOX2, NOTCH1, MIR126 and MIR602.
Similar content being viewed by others
Introduction
The genetic heterogeneity of nonsyndromic recessive deafness (DFNB) reflects the complexity of structure and function of the vertebrate inner ear. Fifty-three DFNB loci have been mapped and reported.1, 2 The rate at which novel DFNB loci have been mapped over the past 10 years has been relatively constant. Approximately five new DFNB loci have been reported each year since 1998 despite the frequent assignment of linkage of deafness segregating families to known DFNB loci. Thus, it has become necessary to ascertain more and more large families to identify a novel DFNB locus. To date, a gene has yet to be identified for 60% of the DFNB loci.
Medlej-Hashim et al.3 reported the mapping of DFNB33 to chromosome 9q34.3 in a single family from Jordan with a LOD score of 3.38. However, the linkage of DFNB33 deafness segregating in the Jordanian family on 9q34.3 was recently reported to be spurious, and subsequently DFNB33 was reassigned to chromosome 10p11.23–q21.1.4 Here, we report three Pakistani families with prelingual hearing loss supporting the existence of a DFNB locus, DFNB79, on chromosome 9q34.3.
Materials and methods
Study participants
This study was approved by the Institutional Review Board (IRB) at the National Centre of Excellence in Molecular Biology (NCEMB), Lahore, Pakistan and the Combined Neuroscience IRB at the National Institutes of Health, USA (OH-93–N-016). Written informed consent was obtained from all of the study participants. Hearing was evaluated by pure-tone (air conduction) audiometry at octave frequencies from 250 to 8000 Hz. Vestibular function was evaluated by tandem gait and Romberg testing. The ocular evaluation comprised slit lamp and funduscopic examinations.
Genotyping and sequencing
Blood was obtained through venipuncture and genomic DNA was extracted using a standard protocol.5 We initially excluded linkage to the reported DFNB loci in the year 2001 using STR (short tandem repeat) markers. Subsequently, a genome-wide scan was undertaken by using 388 fluorescently labeled microsatellite markers spaced at an average interval of 10 cM across the genome (ABI Prism Linkage Mapping Set, v2.5 Applied Biosystems, Foster City, CA, USA). Markers were amplified by polymerase chain reaction (PCR) on a Gene Amp PCR 9700 (Applied Biosystems) and were analyzed on an ABI Prism 3100 Genetic Analyzer. Alleles were assigned using Genescan and Genotyper software (Applied Biosystems). The FASTLINK (http://www.ncbi.nlm.nih.gov/CBBresearch/Schaffer/fastlink.html) computer package was used to calculate LOD scores.6 MLINK was used for two-point LOD scores. Marker order and map distances were obtained from the Marshfield genetic map (http://research.marshfieldclinic.org). An autosomal recessive mode of inheritance with complete penetrance and a disease allele frequency of 0.001 were used for linkage analysis.
Candidate gene screening
Positional candidate genes were identified using the UCSC Genome Bioinformatics web browser (UCSC Genome Bioinformatics Build 36.1: http://genome.ucsc.edu/). Primers used for PCR amplification and sequencing of CACNA1B, EDF1, PTGDS, EHMT1, QSOX2, NOTCH1, MIR126 and MIR602 from the flanking region of each exon were designed using Primer3.
Results
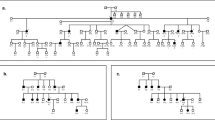
The segregation of hearing loss in families PKDF280, PKDF517 and PKDF741 is consistent with autosomal recessive inheritance (Figures 1 and 2). Affected individuals displayed prelingual, bilateral and severe to profound hearing loss. There is some variability in the severity of hearing loss observed among the affected individuals of DFNB79 families (Figure 4). Only air conduction thresholds were available, and thus we cannot rule out a conductive component of the hearing loss. Tandem gait and Romberg testing were normal in affected individuals indicating that there is no obvious vestibulopathy. We identified no evidence of bone, skin or renal anomalies or a vision disorder. We found no other obvious clinical manifestations co-segregating with hearing loss in the study participants.
Pedigree of family PKDF280. Filled symbols represent hearing-impaired individuals. The linked haplotypes are boxed. The STR markers and their relative map positions (Mb) according to UCSC Genome Bioinformatics Build 36.1 (http://genome.ucsc.edu/) are shown along with the pedigree. Haplotype of PKDF280 defines a linkage region of ∼3.84 Mb. Affected individual VI:2 provided the proximal recombination breakpoint at marker D9S1818 (136.27 Mb).
Pedigrees of families PKDF517 and PKDF741. The linked haplotypes are boxed. Haplotype analysis of family PKDF517 defined a linkage interval of 3.84 Mb bounded by marker D9S1818 (136.27 Mb) and D9SH6 (140.11 Mb). In family PKDF741, the centromeric boundary was established by meiotic recombinations in two affected individuals (IV:4 and IV:5) at D9S1818 (136.27 Mb). The DFNB79 linkage interval in family PKDF741 extends to the telomere.
A preliminary linkage survey of deafness segregating in family PKDF280 revealed no linkage to any known DFNB loci known in 2001. Evidence of linkage to chromosome 9q34.3 was obtained through a subsequent genome-wide linkage analysis. Mapping with additional STR markers and haplotype analysis defined a linkage interval of 3.84 Mb (Figure 1), without a telomeric meiotic breakpoint in PKDF280. However, D9S1818 (136.27 Mb) defined the centromeric boundary in affected individual VI:2 (Figure 1). A significant two-point LOD score (Zmax) of 9.43 at (θ=0) was obtained for the marker D9SH159 (138.33 Mb; Table 1).
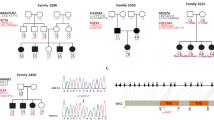
A cohort of more than 700 consanguineous DFNB families has been ascertained in Pakistan at the NCEMB. We screened these families using STR markers and found two additional families segregating deafness (PKDF517 and PKDF741) linked to DFNB79 (Figure 2). PKDF741 yielded a maximum two-point LOD score (Zmax) of 3.25 for markers D9SH159, D9S905, D9SH169 and D9S1838 (Table 1). The maximum two-point LOD score (Zmax) for family PKDF517 was 4.30 at θ=0 for markers D9SH159 and D9SH5 (Table 1). Meiotic recombinations in affected individuals from each DFNB79 family delineate the centromeric boundary at marker D9S1818 (136.27 Mb; Figures 1 and 2). The inferred haplotypes of affected individual V:7 in PKDF517 provided evidence for the telomeric boundary of the DFNB79 locus at marker D9SH6 (Figure 2). Assuming locus homogeneity, these three DFNB79 families define a critical linkage interval of 3.84 Mb on chromosome 9q34.3 (Figure 3), and each of the three DFNB79 families has a distinct haplotype across the refined linkage interval, suggesting different mutant alleles.
DFNB79 chromosomal map location at 9q34.3. Physical map distances are from the UCSC Genome Bioinformatics Build 36.1. STR markers are represented by filled circles. All of the known genes and predicted transcription units in this interval are shown. Candidates are ordered from the proximal gene (RXRA) in column 1 to the distal gene (CR616254) at the bottom of column 4. Markers D9S1818 (136.27 Mb) and D9SH6 (140.11 Mb) define the DFNB79 critical linkage interval.
Three deafness loci have been mapped earlier to chromosome 9, including DFNA36/DFNB7/DFNB117, DFNB318, 9 and DFNA47.10 The genetic interval of DFNB79 does not overlap with any of these three deafness loci (Figure 3). The DFNB79 locus has 113 annotated and hypothetical genes located in an approximately 3.84 Mb interval (Figure 3). The coding exons and flanking intronic sequence of CACNA1B, EDF1, PTGDS, EHMT1, QSOX2, MIR126, MIR602 and NOTOCH1 were sequenced in two affected individuals from each of the three DFNB79 families analyzed in this study, and no pathogenic sequence variants were identified.
Audiograms from an affected individual from each of the three DFNB79 families PKDF280, PKDF517 and PKDF741. The ages at the time of audiological examination are ∼15, 21 and 9 years, respectively. Symbols ‘o’ and ‘x’ denote air conduction pure-tone thresholds at different frequencies in the right and left ear.
Discussion
Families PKDF280, PKDF517 and PKDF741 provide evidence for DFNB79, a novel nonsyndromic recessive deafness locus on chromosome 9q34.3. Each of the three families reported here has a unique DFNB79 haplotype suggesting a multiple allelic series, assuming a single mutated gene is responsible for this phenotype.
There are 113 candidate genes in the DFNB79 critical linkage interval and several of them are expressed in the inner ear,11 including QSOX2, NOTCH1, EDF1, PTGDS, EHMT1 and CACNA1B.12 CACNA1B encodes a voltage-dependent calcium channel. Calcium homeostasis is crucial for mechanoelectrical transduction,13 but no mutations were found in the 46 exons of this gene. A transmembrane receptor, NOTCH1, is crucial for inner ear development.14 However, we did not find a pathogenic mutation in the 34 exons and flanking intronic sequence of NOTCH1 in affected individuals of our DFNB79 families. We also did not find a mutation in EDF1, PTGDS, EHMT1 and QSOX2, which encode a transcription co-activator, an enzyme necessary for the synthesis of prostaglandin, a histone methyltransferase and neuroblastoma-derived sulfhydryl oxidase, respectively. Recently, a dominant mutation of MIR96 responsible for human DFNA50 nonsyndromic deafness15 and a semidominant mutation of Mirn96 causing rapid progressive hearing loss phenotype in the diminuendo mouse16 were reported. The DFNB79 interval includes two microRNAs genes, MIR126 and MIR602, but no mutant alleles were found.
References
Friedman TB, Griffith AJ : Human nonsyndromic sensorineural deafness. Annu Rev Genomics Hum Genet 2003; 4: 341–402.
Morton CC, Nance WE : Newborn hearing screening—a silent revolution. N Engl J Med 2006; 354: 2151–2164.
Medlej-Hashim M, Mustapha M, Chouery E et al: Non-syndromic recessive deafness in Jordan: mapping of a new locus to chromosome 9q34.3 and prevalence of DFNB1 mutations. Eur J Hum Genet 2002; 10: 391–394.
Belguith H, Masmoudi S, Medlej-Hashim M et al: Re-assigning the DFNB33 locus to chromosome 10p11.23-q21.1. Eur J Hum Genet 2009; 17: 122–124.
Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A : A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res 1989; 17: 8390.
Schaffer AA : Faster linkage analysis computations for pedigrees with loops or unused alleles. Hum Hered 1996; 46: 226–235.
Kurima K, Peters LM, Yang Y et al: Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet 2002; 30: 277–284.
Mburu P, Mustapha M, Varela A et al: Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet 2003; 34: 421–428.
Mustapha M, Chouery E, Chardenoux S et al: DFNB31, a recessive form of sensorineural hearing loss, maps to chromosome 9q32-34. Eur J Hum Genet 2002; 10: 210–212.
D'Adamo P, Donaudy F, D'Eustacchio A, Di Iorio E, Melchionda S, Gasparini P : A new locus (DFNA47) for autosomal dominant non-syndromic inherited hearing loss maps to 9p21-22 in a large Italian family. Eur J Hum Genet 2003; 11: 121–124.
Peters LM, Belyantseva IA, Lagziel A, Battey JF, Friedman TB, Morell RJ : Signatures from tissue-specific MPSS libraries identify transcripts preferentially expressed in the mouse inner ear. Genomics 2007; 89: 197–206.
Fettiplace R, Hackney CM : The sensory and motor roles of auditory hair cells. Nat Rev Neurosci 2006; 7: 19–29.
Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K : Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deafwaddler mice. Nat Genet 1998; 19: 390–394.
Lanford PJ, Kelley MW : Notch signaling and cell fate determination in the vertebrate inner ear. Chapter 3 in development of the inner ear. in Kelley MW, Wu DK, Popper AN, Fay RR (eds): Springer Handbook of Auditory Research. Springer: New York, 2005. vol 26, pp 122–157.
Mencia A, Modamio-Hoybjor S, Redshaw N et al: Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet 2009; 41: 609–613.
Lewis MA, Quint E, Glazier AM et al: An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet 2009; 41: 614–618.
Acknowledgements
We are grateful to the participants of this study. We also thank Drs Melanie Barzik, Tamar Ben-Yosef, Byung Yoon Choi, and Dennis Drayna for suggestions regarding this article. We also thank Barbara Ploplis and Erich Boger for their technical assistance. This study was supported by the Higher Education Commission, Islamabad, Pakistan; EMRO/WHO-COMSTECH (RABGH 06–07_24), Ministry of Science and Technology (MoST), Islamabad, and intramural funds from the National Institute on Deafness and Other Communication Disorders, NIH (1 ZO1 DC000039-12) to TBF and (ZO1 DC000064) to AJG.
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic database information
Hereditary Hearing Loss Homepage: http://www.uia.ac.be/dnalab/hhh/
Primer3 Web-Based Server: http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi
UCSC Genome Bioinformatics: http://genome.ucsc.edu/
Morton Fetal Cochlea cDNA Library: http://www.brighamandwomens.org/bwh_hearing/InnerEarcDNAArrays.aspx
NEIBank (select cochlear library): http://neiblast.nei.nih.gov/
Rights and permissions
About this article
Cite this article
Khan, S., Riazuddin, S., Shahzad, M. et al. DFNB79: reincarnation of a nonsyndromic deafness locus on chromosome 9q34.3. Eur J Hum Genet 18, 125–129 (2010). https://doi.org/10.1038/ejhg.2009.121
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.121
Keywords
This article is cited by
-
Identification of autosomal recessive nonsyndromic hearing impairment genes through the study of consanguineous and non-consanguineous families: past, present, and future
Human Genetics (2022)
-
A combined genome-wide association and molecular study of age-related hearing loss in H. sapiens
BMC Medicine (2021)
-
Tprn is essential for the integrity of stereociliary rootlet in cochlear hair cells in mice
Frontiers of Medicine (2019)
-
Haplotype analysis of DFNB8/10 locus reveals contribution of TMPRSS3 mutations in Pakistani deaf population
Genes & Genomics (2014)