Abstract
UHMK1 has previously been implicated as a susceptibility gene for schizophrenia in the 1q23.3 region by significant evidence of allelic and haplotypic association between schizophrenia and several genetic markers at UHMK1 in a London-based case–control sample. Further fine mapping of the UHMK1 gene locus in the University College London schizophrenia case–control sample was carried out with tagging SNPs. Two additional SNPs were found to be associated with schizophrenia (rs6604863 P=0.02, rs10753578 P=0.017). Tests of allelic and haplotypic association were then carried out in a second independent sample from Aberdeen consisting of 858 individuals with schizophrenia and 591 controls. Two of these SNPs also showed association in the Aberdeen sample (rs7513662 P=0.0087, rs10753578 P=0.022) and several haplotypes were associated (global permutation P=0.0004). When the UCL and Aberdeen samples were combined three SNPs (rs7513662 P=0.0007, rs6427680 P=0.0252, rs6694863 P=0.015) and several haplotypes showed association (eg HAP-A, HAP-B, HAP-C permutation P=0.00005). The finding of allelic association with markers in the UHMK1 gene might help explain why it has not been possible, despite great effort, to satisfactorily confirm previously reported associations between schizophrenia and the genes RGS4 and NOS1AP/CAPON. These genes flank UHMK1 and all three loci are within a 700 kb region showing linkage to schizophrenia. The confirmation of association between UHMK1 and schizophrenia, rather than RGS4 and NOS1AP in the London sample, points to the possibility that previous efforts to accurately fine map a gene in the 1q23.3 region have lacked accuracy or may have suffered from methodological flaws.
Similar content being viewed by others
Introduction
Two linkage studies have found significant evidence for linkage between genetic markers on chromosome 1q23.3 and schizophrenia with lods of 6.48 and 3.2.1, 2 Another study from Taiwan found a lod of 0.85 at 1q23.3 over the marker D1S1679 which is between the genes NOS1AP (CAPON) and RGS4.3 A combined allelic association, cytogenetic and family linkage study on chromosome 1 using heterochromatic C-band variants in the 1q22.1–23 region and found cosegregation of a 1qH (C-band) variant and Duffy blood group alleles with schizophrenia in a single family,4 a meta analysis conducted by Lewis et al5 found consistency in linkage studies on 1p13.3–1q23.3 with a significance for non random positive lod scores of P=0.0235 and P=0.0136 under weighted analysis. Other linkage studies from Finland found significant maximal evidence for linkage to schizophrenia with the marker D1S2891 with a lod of 3.8 at 1q32.2 in a position that was telomeric to 1q23.3 but proximal to the DISC1 locus at 1q42.1 DISC 1 locus.6, 7 Two further analyses from Finland in independent family samples found a lod of 3.218 and then 2.7 at the more distal position of 1q42 near the DISC 1 locus.7 Three further linkage studies found less conclusive but supportive evidence for linkage to schizophrenia at 1q23.3 with a lod of 1.7 at a position 20 cM proximal to DISC1.9 A linkage study based on a combined UK/USA sample supported linkage at 1q23.3 with a lod of 1.80 with the marker D1S196, which is the same marker that showed a lod of 3.20 at 1q23.3 in the UK/Icelandic genome scan.2, 10 Any interpretation of these positive 1q23.3 linkage studies must lead to the conclusion that locus heterogeneity is present. However both the family-based parametric lod method and the sib pair linkage method have been employed and both have given positive evidence of linkage in at least two regions of 1q. The lod method in families would tend to favour detection of relatively rare high penetrance loci whereas the sib pair method linkage might be better at detecting common susceptibility variants. At present the linkage studies lead one to strongly suspect that there are two independent schizophrenia susceptibility loci at 1q23.3 and 1q42 but it is uncertain if the disease alleles are at high or low frequency.
Claims that the CAPON and RGS4 genes which are 700 kb apart on chromosome 1q23.3 show genetic association with schizophrenia, have been published but no clear cut replication has been published for any of the markers originally showing association within NOS1AP/CAPON.11, 12, 13 RGS4 has been found to be associated with schizophrenia in a minority of studies14, 15 and some doubt exists about its validity as a schizophrenia susceptibility gene. Our original genetic association studies excluded both the NOS1AP/CAPON and RGS4 loci in the UCL case–control sample.13, 14 Our original study of UHMK116 found positive allelic association with two neighbouring microsatellites (D1Z12 and D1Z13) and two SNPs within UHMK1 (rs10494370 and rs7513662), with two additional SNPs showing a trend towards association (rs423227 and rs6427680). A third microsatellite (D1Z18) in between the steroid dehydrogenase gene HSD17B7 and RGS4 was also weakly associated. The marker D1S1679, which lies between UHMK1 and CAPON has been implicated in two family-based studies of schizophrenia using tests of transmission disequilibrium17 and the extended transmission disequilibrium test (ETDT).18 The ETDT was carried out in multiply affected families, in which situation it cannot distinguish between linkage and association, so the latter finding may just reflect the presence of linkage to 1q23.3. However, the former study does support the presence of association between D1S1679 and schizophrenia and hence that variants in either CAPON or UHMK1, or in both, may influence susceptibility.
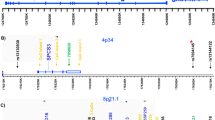
The microsatellite marker D1Z18 is close to the hydroxysteroid (17-β) dehydrogenase 7 gene (HSD17B7) on 1q23.3. We have now genotyped tagging SNPs derived from HapMap to test whether HSD17B7 may be involved in the susceptibility to schizophrenia and have also genotyped additional HapMap tagging SNPs in the UHMK1 gene selected to cover a larger region. We have carried out linkage disequilibrium (LD) analyses of the whole 700 kb region including markers in NOS1AP (CAPON), UHMK1, RGS4 and HSD17B7. Markers localised within UHMK1 and known to show association in the UCL sample have also been genotyped in a replication case–control sample from Aberdeen.
Materials and methods
The methods and DNA samples employed have been described previously in published research. The UCL sample was used to detect genetic association between schizophrenia and the Enthoprotin (Epsin 4) gene19 as well as the Pericentriolar Material 1 gene.20 Part of the Aberdeen sample was employed in a positive replication of association between NRG1 and schizophrenia.21 The UCL case–control sample consisted of DNA from 450 volunteers with schizophrenia (70% male, 30% female, mean age 44.8) and 450 controls (46% male, 54% female, mean age 36.5). All subjects were included only if both parents were of English, Irish, Welsh or Scottish descent and if three out of four grandparents were of the same descent. One grandparent was permitted to be of Caucasian European origin but not of Jewish or non-EU ancestry, based on the EU countries before the recent enlargement (2004). The ancestry information was recorded in an Ancestry Questionnaire and it was confirmed from the family histories recorded in medical records. UK National Health Service (NHS) multicentre and local research ethics committee approval was obtained and all subjects signed an approved consent form after reading an information sheet. All 450 schizophrenic research subjects had previously been diagnosed and assessed by NHS psychiatrists as part of routine clinical diagnosis and treatment. Subjects with short-term drug-induced psychoses, learning disabilities, head injuries and other symptomatic psychoses were excluded. Schizophrenic subjects were recruited on the basis of having an International Classification of Diseases version 10 (ICD10) diagnosis of schizophrenia recorded in the medical case notes. The Schizophrenia and Affective Disorders Schedule-Lifetime Schedule (SADS-L) interview22 was then completed for all cases and controls. Cases were defined as having schizophrenia according to the ‘probable’ level of the Research Diagnostic Criteria (RDC). The ‘supernormal’ control subjects were selected based on not having a family history of schizophrenia, alcoholism or bipolar disorder and for having no past or present personal history of any RDC defined mental disorder.
The replication study sample from Aberdeen consisted of DNA from 858 cases of schizophrenia and 591 controls. The cases were diagnosed at interview with the SCID (N=717) and by use of case note inspection and also by using the OPCRIT to derive DSM IIIR diagnoses. The controls were drawn from the same Scottish population. They were recruited as volunteers via general practices and screened for absence of psychiatric illness. They were sex matched. Genomic DNA was extracted from whole blood samples using a standard cell lysis, proteinase K digestion, phenol/chloroform ethanol precipitation method. All DNA samples were quantified by fluorimetry with Picogreen (Molecular Probes). Tagging SNPs where ascertained for all genes from the international HapMap Project CEU data with the use of the Haploview Tagger function on its default settings.23, 24 In Puri et al16 UHMK1 tagging SNPs covering the genomic extent and 5 Kb upstream and 6.5 Kb downstream region from HapMap phase II Release 19, captured 100% of alleles at r2>0.8 mean max r2=0.972 were used. In this study three additional SNPs were selected and genotyped to further fine map 30 Kb of the downstream region. Nine SNPs within HSD17B7 from HapMap phase II release 20 were selected using Tagger. Eight of these assays were successful and these captured 90% of alleles at r2>0.8 mean max r2=0.996. The assay that failed (rs7530544), had a HapMap CEU minor allele frequency of 0.008. All SNP markers were genotyped by the KASPar method, which is a modification of the Amplifluor method (KBiosciences, Hoddesdon, UK). In all 17% of samples on each microtitre plate were reduplicated to detect error and confirm the reproducibility of genotypes. These data were then analysed to confirm Hardy–Weinberg equilibrium (HWE). Markers with lack of HWE in the control group were rejected and repeated. The genotype data was analysed separately by 96-well DNA plate using SCANGROUP to test for plate by plate differences in haplotype frequencies.25 Some genotyping errors can show up as rare haplotypes occurring on just a single plate whereas true haplotypic associations will be manifest as certain haplotypes being preponderant among cases or controls spread across a number of plates.
Genotypic and allelic associations of individual markers were tested for using χ2 tests. Tests for haplotypic association were carried out using GENECOUNTING, which computes maximum likelihood estimates of haplotype frequencies from phase-unknown data, and the empirical significance was obtained using permutation tests.25, 26, 27 GENECOUNTING was also used to calculate pairwise linkage disequilibrium between all markers, and their relationship was viewed on LocusView 2.0 (Broad Institute).28
Results
The SNPs at HSD17B7 (Figure 1) did not demonstrate allelic or haplotypic association with schizophrenia (Table 1). Results from typing additional markers around UHMK1, along with results from the five SNPs previously found to be associated with schizophrenia16 are shown in Table 2. Two of the new SNPs, rs6604863 and rs10753578, were found to be associated with schizophrenia (P=0.02 and P=0.017 respectively). A third marker, rs6704428, showed a trend towards allelic association (P=0.089). A seven marker haplotype including the original SNPs described in Puri et al16 and the new SNPs described here produced an empirical significance of P=0.0033 (Table 3). These findings from the UCL sample provide additional evidence for UHMK1 as a schizophrenia susceptibility gene.
The following markers were genotyped in the Aberdeen sample: rs10494370, rs7513662, rs423227, rs6427680, rs6604863, rs10753578 and rs6704428. Two of these SNPs yielded evidence for association, rs7513662 (P=0.0087) and rs10753578 (P=0.022) (Table 2). Allele A of rs7513662 was associated with schizophrenia in both the UCL and Aberdeen samples. However, the distribution of allele frequencies for SNP rs10753578 is in the opposite direction to that found in the UCL sample. This phenomenon has been observed before in disease association studies and labelled a ‘Flip-flop’ association.29 It may indicate that the variety of aetiological base pair changes in UHMK1 and the haplotypes they are found within are different in Scotland compared to England.29 However, until such time as the flip flop effect becomes widely accepted as evidence of association in replication studies then our finding in the Aberdeen sample for rs10753578 could also be interpreted as a lack of replication for that SNP. The four SNPs originally implicated in the UCL sample produced evidence for haplotypic association in the Aberdeen sample (empirical permutation P=0.0135) and the presence of two associated SNPs in the Aberdeen sample confirms UHMK1 as a gene increasing susceptibility to schizophrenia. Of relevance is also the fact that haplotypic association with the original four marker haplotype (rs10494370, rs7513662, rs423227 and rs6427680)16 has been confirmed and replicated (permutation P=0.0135), in addition a haplotype analysis of all seven markers in the Aberdeen sample was replicated producing an empirical P-value of 0.0004. Among the other haplotypes tested with less than seven markers, the most significant in the Aberdeen sample comprised markers rs7513662 and rs10753578 (permutation P=0.0002; see Table 3). It should be noted that GENECOUNTING computes significance for all possible haplotypes as a global test and does not give significance for individual haplotypes for a given set of markers.
Data from the UCL and Aberdeen samples were combined and analysed together. Evidence for association was stronger in the combined sample for three SNPs, rs7513662, rs6427680 and rs6694863 (P=0.0007, 0.0252 and 0.015 respectively). Haplotype association for the original four markers was also significant (empirical P=0.011). However, the strongest evidence for association came from haplotype analysis of all seven markers (empirical P=0.00005) as shown in Table 3.
Global haplotypic analyses of the UCL and Aberdeen samples data showed that the same marker loci were associated in both samples. The global analyses carried out by GENECOUNTING include all combinations of alleles for a given set of marker loci and test for both increases and decreases in cases vs controls. However, because of their much greater interest we show only haplotypes that are increased in cases in Table 3. However, inspection of the specific subgroups of alleles at these loci showed some differences between the samples. In the UCL sample, the four locus SNP haplotypes consisting of rs10494370-rs7513662-rs423227-rs6427680 (haplotypes HAP-F and HAP-G) were elevated in cases relative to controls (HAP-F 16.2% cases vs 13% controls; HAP-G 10.2% cases vs 6.5% controls). In the Aberdeen sample, haplotypes HAP-D and HAP-E were elevated in cases relative to controls (32.7% cases and 29.1% controls; 13.8% cases and 11.4% controls respectively). However, in the analysis of the combined samples all four haplotypes were elevated in cases relative to controls (see Table 3). For the 7 SNP haplotype analysis (rs10494370-rs7513662-rs423227-rs6427680-rs6694863-rs10753578-rs6704428) the elevated haplotypes in the UCL sample were HAP-B (12.2% cases 11.7% controls,) and HAP-C (12.8% cases and 10.9% controls). In the Aberdeen sample elevated haplotypes were HAP-A (30.2% cases and 24.8% controls) and HAP-B (13.8% cases and 11.4% controls). Haplotype HAP-B is elevated in both individual samples. However, in the combined analysis all three haplotypes (HAP-A, HAP-B and HAP-C) are more common in cases.
Discussion
The failure to detect association between markers in HSD17B7 and schizophrenia suggests that this gene is unlikely to be involved in schizophrenia susceptibility in the British population. The weakly positive result from D1Z18 could be attributed to detecting LD with aetiological base pair changes in the adjacent UHMK1 gene. Because the markers found to show positive association in the Aberdeen sample had already been shown to be positive in the previously published UHMK1 association study based on the UCL sample we argue on grounds of prior evidence that further correction and re-interpretation of the significance values of the current study are not required. However, after applying a Bonferroni correction (P=0.007 α 0.05) several of the single marker and haplotypic tests of association in the whole sample survive correction for multiple markers. The markers used in the haplotype analyses were overlapping and showed strong intermarker LD. Therefore each new set of haplotypic analysis should be regarded as confirmatory rather than as new evidence for association. We have replicated association between UHMK1 markers and schizophrenia in a second case–control sample. However, the new data from the Aberdeen sample also need to be explained by invoking the flip-flop phenomenon for marker rs10753578. An alternative explanation for this marker is a non-replication. There is a theoretical basis for this phenomenon which has been shown with genetic modeling,29 but we also argue that the conclusions of the study do not depend on this marker alone and that for the other markers the strength of associations with schizophrenia in the combined sample is greater than in either sample alone. As noted above two previous studies have implicated the microsatellite D1S1679 by showing association or linkage with schizophrenia.17, 18 D1S1679 is 105.9 kb from the 5′ end of UHMK1 and displays significant LD (LDPAIRS P-value from Cramer's V) with markers rs7532188, D1Z14, rs164160, rs164167, rs164168 and rs164171 with P-values ⩽1 × 10−5 in the promoter region of UHMK1. It should be noted here that the more familiar D′ and r2 statistics cannot be used for measure of LD with multiallelic microsatellite markers. Evidence for association between schizophrenia and D1S1679 is most likely due to LD with aetiological base pair changes actually being in UHMK1. The fact that the associations reported in the original study and the current replication are not statistically very strong points to the fact that there is locus heterogeneity and also allelic heterogeneity for the involvement of UHMK1 in schizophrenia. We also observed allele and haplotype differences between the two control groups in London and Scotland. This may be due to the fact that we screened out all mental illness in the UCL control sample, which was not done for the Aberdeen control sample. It could also be due to gene allele frequency differences between South England and Scotland, which are very well known to exist. Because both samples had regionally matched controls we do not think any gene frequency differences between London and Scotland would be confounded with evidence for association. It seems most likely that any disease alleles are at low frequency in the populations of South England and Scotland or are of very low penetrance if they are present at moderate or high frequency.
UHMK1 (U2AF homology motif kinase 1) is a serine/threonine-protein kinase with an RNA recognition site and it has been shown to be highly expressed in the brain.30 Both the gene and its protein are highly conserved between mouse, rat and human. UHMK1 is also known to be highly expressed in most parts of the mouse brain, particularly in the amygdala and hippocampus, according to the Allen Brain Atlas. The protein has the ability to phosphorylate classical in vitro substrates such as myelin basic protein and synapsin 131 as well as stathmin, a complex signal relay protein. Stathmin is phosphorylated in response to many signals such as hormone growth and differentiation factors, neurotransmitters or upon activation of T lymphocytes. It has also been proposed as a key regulator of microtubule dynamics.31 UHMK1 was formerly named as KIST (kinase interacting with stathmin) and had originally been discovered by a yeast two hybrid screen using stathmin as bait.32 The stathmin gene locus has been knocked out in the mouse, and it was found that mice homozygous for stathmin gene deletion lacked instinctive fear and had other behavioural abnormalities. The stathmin knockout also had weak memories for past aversive experiences such as those in fear conditioning tests.33 The gene is highly enriched in the lateral nucleus of the amygdala. Because of the known interaction between UHMK1 protein and the stathmin protein one could speculate a knockout of UHMK1 would also produce similar behavioural abnormalities. A study comparing anterior cingulate cortex grey matter proteomes between schizophrenia and controls found that stathmin was increased by 1.8 fold change in a schizophrenia cohort.34 It has also been found that the expression of UHMK1 is significantly downregulated in mice treated with the antipsychotic drugs clozapine and haloperidol (Rizig et al 2008 (submitted)). Further independent replications of the association between UHMK1 and schizophrenia are now needed in as many populations as possible. Re-sequencing, gene expression analysis and several types of conditional and knockout transgenic mice can now be created to explore the normal and abnormal biological functions of the UHMK1 gene.
References
Brzustowicz LM, Hodgkinson KA, Chow EW, Honer WG, Bassett AS : Location of a major susceptibility locus for familial schizophrenia on chromosome 1q21-q22. Science 2000; 288: 678–682.
Gurling HM, Kalsi G, Brynjolfson J et al: Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 2001; 68: 661–673.
Hwu HG, Liu CM, Fann CS, Ou-Yang WC, Lee SF : Linkage of schizophrenia with chromosome 1q loci in Taiwanese families. Mol Psychiatry 2003; 8: 445–452.
Kosower NS, Gerad L, Goldstein M et al: Constitutive heterochromatin of chromosome 1 and Duffy blood group alleles in schizophrenia. Am J Med Genet 1995; 60: 133–138.
Lewis CM, Levinson DF, Wise LH et al: Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: Schizophrenia. Am J Hum Genet 2003; 73: 34–48.
Hovatta I, Varilo T, Suvisaari J et al: A genomewide screen for schizophrenia genes in an isolated Finnish subpopulation suggesting multiple susceptibility loci. Am J Hum Genet 1999; 65: 1114–1124.
Ekelund J, Hennah W, Hiekkalinna T et al: Replication of 1q42 linkage in Finnish schizophrenia pedigrees. Mol Psychiatry 2004; 9: 1037–1041.
Ekelund J, Lichtermann D, Hovatta I et al: Genome-wide scan for schizophrenia in the Finnish population: evidence for a locus on chromosome 7q22. Hum Mol Genet 2000; 9: 1049–1057.
Cai G, Wu X, Li T et al: Linkage analysis of susceptibility genes for familial schizophrenia on chromosome 1 in Chinese population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2002; 19: 491–494.
Shaw SH, Kelly M, Smith AB et al: A genome-wide search for schizophrenia susceptibility genes. Am J Med Genet 1998; 81: 364–376.
Brzustowicz LM, Simone J, Mohseni P et al: Linkage disequilibrium mapping of schizophrenia susceptibility to the CAPON region of chromosome 1q22. Am J Hum Genet 2004; 74: 1057–1063.
Zheng Y, Li H, Qin W et al: Association of the carboxyl-terminal PDZ ligand of neuronal nitric oxide synthase gene with schizophrenia in the Chinese Han population. Biochem Biophys Res Commun 2005; 328: 809–815.
Puri V, McQuillin A, Thirumalai S et al: Failure to confirm allelic association between markers at the CAPON gene locus and schizophrenia in a British sample. Biol Psychiatry 2006; 59: 195–197.
Rizig MA, McQuillin A, Puri V et al: Failure to confirm genetic association between schizophrenia and markers on chromosome 1q23.3 in the region of the gene encoding the regulator of G-protein signalling 4 protein (RGS4). Am J Med Genet, Neuropsychiatr Genet 2006; 141: 296–300.
Talkowski ME, Seltman H, Bassett AS et al: Evaluation of a susceptibility gene for schizophrenia: genotype based meta-analysis of RGS4 polymorphisms from thirteen independent samples. Biol Psychiatry 2006; 60: 152–162.
Puri V, McQuillin A, Choudhury K et al: Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry 2007; 61: 873–879.
Miranda A, Garcia J, Lopez C et al: Putative association of the carboxy-terminal PDZ ligand of neuronal nitric oxide synthase gene (CAPON) with schizophrenia in a Colombian population. Schizophr Res 2006; 82: 283–285.
Rosa A, Fananas L, Cuesta MJ, Peralta V, Sham P : 1q21-q22 locus is associated with susceptibility to the reality-distortion syndrome of schizophrenia spectrum disorders. Am J Med Genet 2002; 114: 516–518.
Pimm J, McQuillin A, Thirumalai S et al: The Epsin 4 gene on chromosome 5q, which encodes the clathrin-associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am J Hum Genet 2005; 76: 902–907.
Gurling HM, Critchley H, Datta SR et al: Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiatry 2006; 63: 844–854.
Stefansson H, Sarginson J, Kong A et al: Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet 2003; 72: 83–87.
Spitzer RL, Endicott J : The Schedule for Affective Disorders and Schizophrenia, Lifetime Version, 3rd edn New York: New York State Psychiatric Institute, 1977.
Barrett JC, Fry B, Maller J, Daly MJ : Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263–265.
de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D : Efficiency and power in genetic association studies. Nat Genet 2005; 37: 1217–1223.
Curtis D, Knight J, Sham PC : Program report: GENECOUNTING support programs. Ann Hum Genet 2006; 70: 277–279.
Zhao JH, Curtis D, Sham PC : Model-free analysis and permutation tests for allelic associations. Hum Hered 2000; 50: 133–139.
Zhao JH, Lissarrague S, Essioux L, Sham PC : GENECOUNTING: haplotype analysis with missing genotypes. Bioinformatics 2002; 18: 1694–1695.
Petryshen T, Kirby A, Ainscow M : LocusView ‘unpublished software’ from Broad Institute 2003.URL: www.broad.mit.edu/mpg/locusview/.
Lin PI, Vance JM, Pericak-Vance MA, Martin ER : No gene is an island: the flip-flop phenomenon. Am J Hum Genet 2007; 80: 531–538.
Bieche I, Manceau V, Curmi PA et al: Quantitative RT-PCR reveals a ubiquitous but preferentially neural expression of the KIS gene in rat and human. Brain Res Mol Brain Res 2003; 114: 55–64.
Maucuer A, Ozon S, Manceau V et al: KIS is a protein kinase with an RNA recognition motif. J Biol Chem 1997; 272: 23151–23156.
Maucuer A, Camonis JH, Sobel A : Stathmin interaction with a putative kinase and coiled-coil-forming protein domains. Proc Natl Acad Sci USA 1995; 92: 3100–3104.
Shumyatsky GP, Malleret G, Shin RM et al: stathmin, a gene enriched in the amygdala, controls both learned and innate fear. Cell 2005; 123: 697–709.
Clark D, Dedova I, Cordwell S, Matsumoto I : A proteome analysis of the anterior cingulate cortex gray matter in schizophrenia. Mol Psychiatry 2006; 11: 459–470, 423.
Acknowledgements
This research was funded by the Neuroscience Research Charitable Trust. Sample collection in Scotland was funded by University of Aberdeen and Glaxo Smith Kline PLC. The authors have no commercial conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Electronic References:
International HapMap Project http://www.hapmap.org/
Haploview http://www.broad.mit.edu/mpg/haploview/download.php
KBiosciences http://www.kbioscience.co.uk
Allen Brain Atlas http://www.brainatlas.org/aba/
LocusView http://www.broad.mit.edu/mpg/locusview/
Rights and permissions
About this article
Cite this article
Puri, V., McQuillin, A., Datta, S. et al. Confirmation of the genetic association between the U2AF homology motif (UHM) kinase 1 (UHMK1) gene and schizophrenia on chromosome 1q23.3. Eur J Hum Genet 16, 1275–1282 (2008). https://doi.org/10.1038/ejhg.2008.76
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.76