Abstract
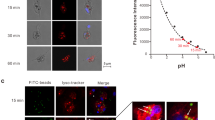
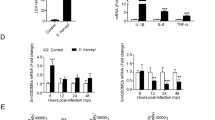
High-mobility group box (HMGB) proteins, a family of chromatin-associated nuclear proteins, play amazingly multifaceted roles in the immune system of mammals. Thus far, little is known about the nucleocytoplasmic distribution of HMGBs in teleosts. The present study systematically investigated the dynamic localization of all six HMGB proteins in Ctenopharyngodon idella kidney (CIK) cells. Under basal conditions, all HMGBs exclusively localized to the nucleus. Grass carp reovirus (GCRV), polyinosinic–polycytidylic (poly(I∶C)) potassium salt and lipopolysaccharide (LPS) challenge evoked the nuclear export of HMGBs to various degrees: GCRV challenge induced the highest nuclear export of CiHMGB2b, and poly(I∶C) and LPS evoked the highest nucleocytoplasmic shuttling of CiHMGB1b. Overall, the nucleocytoplasmic shuttling of CiHMGB2a and CiHMGB3b was rarely induced by these challenges. Dynamic imaging uncovered that the nucleocytoplasmic GCRV-induced relocation of CiHMGB2b occurred in cells undergoing karyotheca rupture, apoptosis or proliferation. Western blot analyses were used to examine HMGB-EGFP fusion proteins in whole cell lysates, cytosol, nuclear fractions and culture medium. Further investigation demonstrated the nuclear retention of N-terminal HMG-boxes and the nucleocytoplasmic distribution of the C-terminal acidic tails. Comparative analyses of the dynamic relocation of full-length, truncated or chimeric HMGBs confirmed that the intramolecular interaction between HMG-boxes and C-tail domains mediated the nucleocytoplasmic translocation of HMGBs. These results not only provide an overall understanding of the subcellular localization of HMGBs, but also reveal the induction mechanism of the nucleocytoplasmic translocation of HMGBs by GCRV challenge, which lays a foundation for further studies on the interactions among pathogens, HMGBs and pattern recognition receptors in the innate immunity of teleosts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Stros M . HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta 2010; 1799: 101–113.
Moleri S, Cappellano G, Gaudenzi G, Cermenati S, Cotelli F, Horner D et al. The HMGB protein gene family in zebrafish: evolution and embryonic expression patterns. Gene Expr Patterns 2011; 11: 3–11.
Yang C, Peng L, Su J . Two HMGB1 genes from grass carp Ctenopharyngodon idella mediate immune responses to viral/bacterial PAMPs and GCRV challenge. Dev Comp Immunol 2013; 39: 133–146.
Yang C, Chen L, Su J, Feng X, Rao Y . Two novel homologs of high mobility group box 3 gene in grass carp (Ctenopharyngodon idella): potential roles in innate immune responses. Fish Shellfish Immunol 2013; 35: 1501–1510.
Rao Y, Su J, Yang C, Peng L, Feng X, Li Q . Characterizations of two grass carp Ctenopharyngodon idella HMGB2 genes and potential roles in innate immunity. Dev Comp Immunol 2013; 41: 164–177.
Ueda T, Yoshida M . HMGB proteins and transcriptional regulation. Biochim Biophys Acta 2010; 1799: 114–118.
Yanai H, Ban T, Taniguchi T . High-mobility group box family of proteins: ligand and sensor for innate immunity. Trends Immunol 2012; 33: 633–640.
Gougeon ML, Melki MT, Saidi H . HMGB1, an alarmin promoting HIV dissemination and latency in dendritic cells. Cell Death Differ 2012; 19: 96–106.
Xie J, Hodgkinson JW, Li C, Kovacevic N, Belosevic M . Identification and functional characterization of the goldfish (Carassius auratus L.) high mobility group box 1 (HMGB1) chromatin-binding protein. Dev Comp Immunol 2014; 44: 245–253.
Yanai H, Ban T, Wang Z, Choi MK, Kawamura T, Negishi H et al. HMGB proteins function as universal sentinels for nucleic-acid-mediated innate immune responses. Nature 2009; 462: 99–103.
Bianchi ME, Celona B . Ancient news: HMGBs are universal sentinels. J Mol Cell Biol 2010; 2: 116–117.
Yanai H, Chiba S, Ban T, Nakaima Y, Onoe T, Honda K et al. Suppression of immune responses by nonimmunogenic oligodexynucleotides with HMGBs. Proc Natl Acad Sci USA 2011; 108: 11542–11547.
Moisy D, Avilov SV, Jacob Y, Laoide BM, Ge X, Baudin F et al. HMGB1 protein binds to influenza virus nucleoprotein and promotes viral replication. J Virol 2012; 86: 9122–9133.
Chen Y, Jia X, Huang X, Zhang S, Li M, Xie J et al. Two Litopenaeus vannamei HMGB proteins interact with transcription factors LvSTAT and LvDorsal to activate the promoter of white spot syndrome virus immediate-early gene ie1. Mol Immunol 2011; 48: 793–799.
Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J 2003; 22: 5551–5560.
Müller S, Ronfani L, Bianchi ME . Regulated expression and subcellular localization of HMGB1, a chromatin protein with a cytokine function. J Intern Med 2004; 255: 332–343.
Yang C, Su J, Huang T, Zhang R, Peng L . Identification of a retinoic acid-inducible gene I from grass carp (Ctenopharyngodon idella) and expression analysis in vivo and in vitro. Fish Shellfish Immunol 2011; 30: 936–943.
Wang Q, Zeng W, Liu C, Zhang C, Wang Y, Shi C et al. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J Virol 2012; 86: 12466.
Fan Y, Rao S, Zeng L, Ma J, Zhou Y, Xu J et al. Identification and genomic characterization of a novel fish reovirus, Hubei grass carp disease reovirus, isolated in 2009 in China. J Gen Virol 2013; 94: 2266–2277.
Peng L, Yang C, Su J . Protective roles of grass carp Ctenopharyngodon idella Mx isoforms against grass carp reovirus. PLoS One 2012; 7: e52142.
Kaczmarczyk SJ, Sitaraman K, Hill T, Hartley JL, Chatterjee DK . Tus, an E. coli protein, contains mammalian nuclear targeting and exporting signals. PLoS One 2010; 5: e8889.
Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M et al. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem 2009; 284: 478–485.
Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH . Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem 2007; 282: 5101–5105.
Sessa L, Bianchi ME . The evolution of high mobility group box (HMGB) chromatin proteins in multicellular animals. Gene 2007; 387: 133–140.
Reeves R . Nuclear functions of the HMG proteins. Biochim Biophys Acta 2010; 1799: 3–14.
Chen R, Hou W, Zhang Q, Kang R, Fan XG, Tang D . Emerging role of high-mobility group box 1 (HMGB1) in liver diseases. Mol Med 2013; 19: 357–366.
Cao S, Li S, Li H, Xiong L, Zhou Y, Fan J et al. The potential role of HMGB1 release in peritoneal dialysis-related peritonitis. PLoS One 2013; 8: e54647.
Fan C, Shao L, Fang Q . Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol 2010; 155: 1755–1763.
Shao L, Guo H, Yan LM, Liu H, Fang Q . Aquareovirus NS80 recruits viral proteins to its inclusions, and its C-terminal domain is the primary driving force for viral inclusion formation. PLoS One 2013; 8: e55334.
Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J 2003; 22: 5551–5560.
Barqasho B, Nowak P, Abdurahman S, Walther-Jallow L, Sonnerborg A . Implications of the release of high-mobility group box 1 protein from dying cells during human immunodeficiency virus type 1 infection in vitro. J Gen Virol 2010; 91: 1800–1809.
Andersson U, Tracey K . HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol 2011; 29: 139–162.
Sachdev U, Cui X, McEnaney R, Wang T, Benabou K . TLR2 and TLR4 mediate differential responses to limb ischemia through MyD88-dependent and independent pathways. PLoS One 2012; 7: e50654.
Yang H, Tracey KJ . Targeting HMGB1 in inflammation. Biochim Biophys Acta 2010; 1799: 149–156.
Scaffidi P, Misteli T, Bianchi ME . Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002; 418: 191–195.
Pusterla T, de Marchis F, Palumbo R, Bianchi ME . High mobility group B2 is secreted by myeloid cells and has mitogenic and chemoattractant activities similar to high mobility group B1. Autoimmunity 2009; 42: 308–310.
Ivanov S, Dragoi AM, Wang X, Dallacosta C, Louten J, Musco G et al. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood 2007; 110: 1970–1981.
Chiba S, Baghdadi M, Akiba H, Yoshiyama H, Kinoshita I, Dosaka-Akita H et al. Tumor-infiltrating DCs suppress nucleic acid-mediated innate immune responses through interactions between the receptor TIM-3 and the alarmin HMGB1. Nat Immunol 2012; 13: 832–842.
Yu Y, Xie M, Kang R, Livesey KM, Cao L, Tang D . HMGB1 is a therapeutic target for leukemia. Am J Blood Res 2012; 2: 36–43.
Kang R, Livesey KM, Zeh HJ 3 rd, Lotze MT, Tang D . Metabolic regulation by HMGB1-mediated autophagy and mitophagy. Autophagy 2011; 7: 1256–1258.
Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, van Houten B et al. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 2011; 13: 701–711.
Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene 2014; 33: 567–577.
Taniguchi N, Carames B, Ronfani L, Ulmer U, Komiya S, Bianchi ME et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci USA 2009; 106: 1181–1186.
Suzuki S, Takeshita K, Asamoto M, Takahashi S, Kandori H, Tsujimura K et al. High mobility group box associated with cell proliferation appears to play an important role in hepatocellular carcinogenesis in rats and humans. Toxicology 2009; 255: 160–170.
Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ et al. Overexpression of high-mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res 2010; 16: 5511–5521.
Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J . The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol 2010; 11: 1005–1013.
Matsumoto Y, Hayashi Y, Omori H, Honda T, Daito T, Horie M et al. Bornavirus closely associates and segregates with host chromosomes to ensure persistent intranuclear infection. Cell Host Microbe 2012; 11: 492–503.
Pedersen DS, Merkle T, Marktl B, Lildballe DL, Antosch M, Bergmann T et al. Nucleocytoplasmic distribution of the Arabidopsis chromatin-associated HMGB2/3 and HMGB4 proteins. Plant Physiol 2010; 154: 1831–1841.
Mosevitsky MI, Novitskaya VA, Iogannsen MG, Zabezhinsky MA . Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem 1989; 185: 303–310.
Youn JH, Shin JS . Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J Immunol 2006; 177: 7889–7897.
Schmidt-Zachmann MS, Dargemont C, Kühn LC, Nigg EA . Nuclear export of proteins: the role of nuclear retention. Cell 1993; 74: 493–504.
Shirakawa H, Tanigawa T, Sugiyama S, Kobayashi M, Terashima T, Yoshida K et al. Nuclear accumulation of HMG2 protein is mediated by basic regions interspaced with a long DNA-binding sequence, and retention within the nucleus requires the acidic carboxyl terminus. Biochemistry 1997; 36: 5992–5999.
Acknowledgements
The authors thank Dr Chong Zhang for live cell imaging assistance and Professor Shantin Zhao and Dr Jiutao Wang for confocal microscopy assistance. This study was supported by the Program for New Century Excellent Talents in University (NCET-08-0466) and the Chinese Universities Scientific Fund (QN2009022).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Cellular & Molecular Immunology's website (http://www.nature.com/cmi).
Supplementary information
Rights and permissions
About this article
Cite this article
Rao, Y., Su, J., Yang, C. et al. Dynamic localization and the associated translocation mechanism of HMGBs in response to GCRV challenge in CIK cells. Cell Mol Immunol 12, 342–353 (2015). https://doi.org/10.1038/cmi.2014.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2014.55