Abstract
During mammalian programmed cell death, cleavage of the translation initiation factor 4G proteins (eIF4GI and eIF4GII) by caspase-3 induces the cap-independent synthesis of pro-apoptotic proteins. Apoptosis occurs naturally in the gonad to remove germ cells that are not selected to grow as oocytes and mature into eggs. Here, we describe two major isoforms of Caenorhabditis elegans eIF4G that are derived from a single gene (ifg-1) and their separate roles in germline homeostasis. Full length IFG-1 protein (170 kDa isoform) differs from the shorter isoform (130 kDa) by the inclusion of the N-terminal domain containing the putative eIF4E-binding site required for mRNA cap recognition. Depletion of the cap-associated p170 isoform induced CED-4 expression in oocytes and markedly increased germline apoptotic events, but did not prevent early mitotic germ cell proliferation. Loss of both p170 and p130 suppressed germ cell proliferation and arrested larval development. Evidence suggests that eIF4G isoforms are differentially utilized during oogenesis to regulate germ cell apoptosis. We propose that an alternative mechanism to eIF4G cleavage may be employed in germ cells by changing the availability of the p170 isoform.
Similar content being viewed by others
Main
Translational regulation has come to be recognized as a critical mode of gene regulation in germ cell and embryonic development.1 Translational control reflects not only changes in the overall rate of protein synthesis but also the preferential translational activation of certain mRNAs. During development maternal mRNAs synthesized during oogenesis are inherited by the fertilized egg and translated in the embryo during temporally and spatially defined episodes of mRNA recruitment.2 Coincidentally, translational control and cell proliferation are two cellular processes that are reactivated during the progression of oncogenesis.3 Regulation of mRNA utilization occurs mainly through the activity of translation initiation factors. mRNA recruitment is catalyzed by eIF4E, which binds the 7-methylguanosine cap at the mRNA 5′ end, and its association with eIF4G, a scaffold protein, to catalyze joining of mRNA to the 40S ribosomal subunit.4 Assembly of a stable 48S initiation complex is the committed step for protein synthesis for essentially all mRNAs.
eIF4E and eIF4G are represented by multiple isoforms in all eukaryotic species examined.5 Nematode eIF4E isoforms are expressed in tissue-specific patterns and have specialized roles in those tissues.6,7,8,9,10 Mammalian eIF4GI and eIF4GII are full length isoforms encoded by separate genes that are broadly co-expressed.11 eIF4G contains three modular structural domains. The N-terminus binds to PABP and eIF4E.4 An exposed hinge region links the N-terminal domain to the conserved central and C-terminal domains, which bind eIF3, eIF4A, and the ribosome. p97/DAP5 is a shorter eIF4G isoform, encoded by a third gene, that contains the central and C-terminal domains but not the N-terminal domain. Consequently, p97 is able to promote cap-independent translation of mRNAs containing an IRES.12 Cellular crises such as viral infection or apoptosis disrupt the balance of cap-dependent and cap-independent initiation. Caspase-3 cleaves eIF4GI, eIF4GII and p97. The central domain products stimulate cap-independent synthesis of the apoptosome protein, Apaf-1, and of p97 itself.13 By this mechanism caspase-3 mediates a global reprogramming of protein synthesis that promotes the final execution phase of programmed cell death.14
The natural purpose for apoptosis as a cell fate is most evident during embryonic development. In animal embryos, lineages of cells must die off for the architecture of adult organ systems to take shape.15 This is even true of some cells in the immortal lineage of the germline.16 Among the metabolic activities that direct oocyte and sperm differentiation, the synthesis of new proteins is perhaps most crucial.6,17,18,19,20 In the present study, we show that C. elegans eIF4G (hereafter referred to as IFG-1) is encoded by a single gene, but exists in two major isoforms that differ in their N-termini and their association with cap-binding complexes. The balance between short and long eIF4G isoforms contributes to a fate decision, specifically between oocyte meiotic progression and apoptosis. Such observations require us to re-think the classical role of translation factors as part of the housekeeping machinery in the cell.
Results
ifg-1 mRNAs and protein isoforms
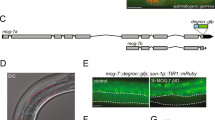
Over thirty expressed sequence tags encoding ifg-1 mRNAs were identified in the WormBase sequence database. The sequences indicate sets of coding mRNAs that differ in their 5′ start sites. One set contains the 5′-most exons 1 through 4, while the second begins near the end of exon 4, approximately a third of the way through the ifg-1 gene (Figure 1). mRNAs of approximately 4500 and 2500 nt were detected by northern blot analysis from wild type C. elegans, correlating well with the EST predictions. More precise mapping of the mRNA 5′ ends was accomplished by RNase protection assay. The most abundant protected fragment (‘a’) indicates an mRNA start site at the end of exon 4 and corresponds to the ‘short’ mRNA (2500 nt) detected by northern blot. The start site for this shorter mRNA precedes several in-frame AUG start codons in exons 5 and 6. This mRNA is predicted to encode p130 (see Figure 2) containing the MIF4G-conserved region.23 Longer protected fragments (1600–2000 nt; ‘b, c’) represent alternatively spliced ifg-1 mRNAs (4500 nt) with end points near the 5′ ends of exons 1–4 (Figure 1). Thus, C. elegans ifg-1 mRNAs encode two significantly different isoforms (p170 and p130) that include or exclude, respectively, the N-terminal third of eIF4G.
Multiple mRNAs derived from the C. elegans eIF4G (ifg-1) gene. (a) ifg-1 gene structure, putative mRNAs, and the region of hybridization for the antisense RNase protection probe SX. (Schematic is drawn to scale). Numbered boxes correspond to the nine predicted ifg-1 exons. Splicing of these exons was confirmed by RT-PCR (data not shown). The mRNAs correspond to expressed sequence tags (ESTs) yk1611h10, yk450c12, yk1422b10, and yk249h6. The 5′ cap and polyadenylated tail are included to distinguish processed transcripts from gene. (b) Northern blot hybridization and RNase protection analysis of ifg-1 mRNA expression. Total RNA (and purified poly(A)+ RNA, not shown) from wild type (+/+) and heterozygous ifg-1(ok1211) (−/+) worms analyzed by northern blot revealed ifg-1 mRNAs of approximately 900 nt (−), 2500 nt (a), and 4500 nt (b, c). Sizes were determined relative to 18S and 28S rRNAs as described in Methods. For RNase protection assay, total wild type RNA (Ce) or yeast (Sc) RNA was hybridized to the uniformly labeled 2083 nt antisense RNA probe (P) corresponding to the 5′ half of the longest EST and digested with RNases A and T1 at various dilutions (0, 0.01, 0.004). A larger labeled RNA (∼3000 nt) likely derives from antisense transcription of incompletely linearized plasmid template. The most abundant ifg-1-protected fragment, ‘a’ (740 nt) maps the 5′ end of the short (p130) mRNA to the start of the yk249h6 cDNA. Protected fragment ‘b’ maps near the end of yk450c12, and the ‘c’ protected fragments map to the end of yk1611h10 as well as the sites for introns 1 and 2. The latter fragments arose from an alternative splice of intron 1 that includes four extra codons, as confirmed by RT-PCR and sequencing (data not shown). Micro-heterogeneity exists within the p170 ifg-1 mRNA that gives rise to a variety of closely related N-terminal structures, similar to that found for human eIF4GI.21 The position of non-specific bands (*) were identified in yeast RNA samples
Two major IFG-1 isoforms, p170 and p130, are present in embryos, larvae, and adult worms. (a) Schematic of human and C. elegans eIF4G proteins. Human eIF4GI, -II, and 4G-like p97 are all derived from separate genes. The regions of eIF4G that bind other translation initiation factors are shown as hatched and stippled boxes. Also shown are two major eIF4G (IFG-1) isoforms in C. elegans described in this study, p170 and p130. The central domain highly conserved among all eIF4G-family proteins (MIF4G) is present in both p170 and p130 IFG-1. The antigenic sites for domain-specific antibodies used to detect isoforms of IFG-1 that contain or lack the N-terminal region are shown. (b) ‘N-terminal’ and ‘Central’ domain antibodies detect protein isoforms of IFG-1 in lysates from wild type worms. Lysates were also subject to m7GTP affinity chromatography to determine association with an mRNA cap-binding fraction. Elution fractions were immunoblotted with central domain antibody for both p170 and p130. p170 was present in both the unbound and bound fractions. p130 was present only in the unbound protein fraction. A breakdown product from p170 is also shown in the bound fraction (carrot). (c) IFG-1 p170 and p130 expression in whole wild type (WT) worms and worms devoid of germline (glp-4) showed no enrichment for either isoform in the germline. Worms mutant in the glp-4 gene were devoid of gonadal tissue when grown at 25°C, as evidenced by the lack of the P granule protein, PGL-1, that characterizes germ cells. The relative p170 and p130 abundance in glp-4 worms; however, appeared largely unchanged, as did the abundance of each isoform in pre-hatching embryos (Eggs). Actin was a control for the amount of protein loaded. (d) All larval stages of development through adulthood likewise contained both p170 and p130 in similar proportions. The abundance of both IFG-1 isoforms is lowest during the L2 stage, in agreement with previously reported ifg-1 mRNA abundance22
Both IFG-1 isoforms were detected with similar abundance in whole worm extracts using antisera against the shared central region (Figure 2). Likewise, IFG-1 isoforms were equally distributed in embryos, larvae, adult germline and somatic tissues. However, only p170 IFG-1 was affinity purified on m7GTP-Sepharose, and thus appears to be part of the canonical mRNA cap-binding complex. p130 IFG-1, on the other hand, remained in the unbound fraction, indicating that it does not bind any of the five C. elegans eIF4E isoforms (IFEs) that all have demonstrated affinities to m7GTP.8 This is consistent with the absence of a predicted eIF4E-binding site in p130. Western blotting with an N-terminal antiserum detected p170 but failed to recognize p130. Thus, the IFG-1 isoforms are not likely to be functionally equivalent. Unlike p170, p130 does not tightly associate with any eIF4E isoforms and may be a paralog of the p97 protein, which is encoded by a distinct gene in humans.4,12
Efficient depletion of long and short ifg-1 mRNA by targeted RNAi
To address the role of individual isoforms in the germline, we initially depleted ifg-1 mRNA using RNAi. To minimize off-target effects and identify specific phenotypes for the isoforms, we designed several short RNAi target sequences corresponding to various regions of p170 mRNA. N2 (RNAi) targets only the long ifg-1 mRNA isoform (Figure 3). RT-PCR verified that p170 ifg-1 mRNA was depleted in N2 (RNAi) treated worms relative to actin mRNA. As expected, p130 mRNA remained intact since N2 (RNAi) sequences do not overlap. In contrast, centralized RNAi targets, N1, C2, and C3 depleted both p170 and p130 mRNAs (Figure 3b). We further quantified the extent of p170 ifg-1 mRNA depletion in independent experiments by qRT-PCR. N2, N1 and C3 (RNAi) generally removed 70–80% of mRNA encoding the cap-binding isoform (Figure 3c). Depletion by C2 (RNAi) varied more widely (40–80%), which might explain its variable severity in phenotype (see Figure 4). These data demonstrate a selective knock down of p170 mRNA relative to p130 mRNA by N2 (RNAi).
Selective depletion of p170 and p130 mRNA by RNA interference. Delivery of dsRNA in worms by feeding has been shown to provide a continuous supply of dsRNA to worms over all of larval and adult stages.24 Short dsRNA sequences corresponding to ifg-1 mRNAs encoding p170 or both p170 and p130 were fed to wild type hermaphrodites. (a). Diagram representing p170 and p130 mRNA, the complementary positions of RNAi targets (boxes N2, N1, C2 and C3), and regions of amplification by RT-PCR (double arrows) and qRT-PCR. N2 is complementary to the 5′ end of ifg-1 mRNA (primarily exons 1–3) and therefore depletes only the long ifg-1 mRNA isoform. Centralized RNAi targets, N1 (exon 4-5 junction) and C2 (exon 6–7 junction), and the 3′ target C3 (within exon 7) were designed to deplete both p170 and p130 mRNAs. (b) To determine extent of depletion of full length (p170) ifg-1 mRNA, RT-PCR was conducted on total RNA from offspring of RNAi adults. RNAi-treated ‘mothers’ showed dramatically reduced fertility, typically producing less than 100 offspring, which greatly limited the amount of sample RNA for analysis. Primers amplified sequences at the junction of exons 3 and 4 (Upstream) to determine the remaining level of p170 mRNA. Alternatively, primers that cover the junctions of exons 4, 5 and 6 (Downstream) were used to determine the extent of both p130 and p170 mRNA depletion. Total RNA was extracted from 50 RNAi-treated F1 adults at 72 h by micro-scale RNA isolation and amplified by RT-PCR. Amplicons included an intron–exon boundary to distinguish mRNA products from unspliced or genomic products. Actin primers were also used as an endogenous control. RNA that was not DNase I-treated (VD-) was a control for genomic DNA contamination. (c) Real time PCR (qRT-PCR) was used to quantify ifg-1 p170 mRNA depletion from all five RNAi treatments (V, N2, N1, C2 and C3) in two independent fed RNAi experiments. Triplicate determinations of ifg-1 and GAPDH mRNA Ct values were used to plot relative mRNA abundance calculated by 2−ΔΔCt
Depletion of IFG-1 prevents oocyte development late in meiosis. (a) Diagram of one lobe of an adult hermaphrodite gonad. Each U-shaped lobe contains maturing oocytes that proliferate mitotically in the distal end, progress through meiotic prophase I to diplotene/diakinesis at the bend where chromosome condensation and cellularization occurs. At this point oocytes begin growth and align single file. They move proximally to become fertilized in the spermatheca and are deposited in a common uterus. Natural germ cell apoptotic events occur just prior to diakinesis. (b) DIC images of (top to bottom) vector (V)-, ifg-1 N2-, N1-, C2-, and C3-(RNAi) adult hermaphrodites. Black arrowheads indicate full-grown oocytes in proximal arm. In control RNAi-treated worm gonads (V), immature oocytes produced in the distal gonad matured to neatly packed full grown oocytes in the proximal gonad. These oocytes were efficiently fertilized and produced embryos that accumulated in the uterus. By contrast, few late stage oocytes were present in ifg-1(RNAi) treated worms and few embryos were produced. Sperm nuclei were frequently observed in the spermatheca suggesting that spermatogenesis was less affected than oogenesis by IFG-1 depletion (N2). In general, ifg-1(RNAi) caused approximately 25–30% reduction in size compared to control-treated worms. All ifg-1(RNAi) worms displayed marked deterioration in the proximal gonad (*). The distal gonad is obscured by the gut in some images. The number of worms observed by microscopy were: V (n=23), N2 (n=16), N1 (n=12), C2 (n=28) and C3 (n=14). (c) Fluorescence microscopy of DAPI-stained control (V) and ifg-1(RNAi) hermaphrodites. The mitotic and meiotic nuclei of oocytes developing in the distal gonad are shown (white dotted line). ifg-1 N2-, N1-, and C2-(RNAi) worms did not produce fully grown oocytes as evidenced by the lack of condensed bivalent chromosomes (white arrowheads) in the proximal gonad. The number of worms observed by microscopy were: V (n=7), N2 (n=2), N1 (n=3), C2 (n=2) and C3 (n=6). The data are representative of five independent experiments
ifg-1(RNAi) causes local defects in gametogenesis
As a result of ifg-1(RNAi), F1 worms exhibited a loss of fertility and varied degrees of stunted growth. To determine if infertility was a consequence of defects in germ cell development, we examined RNAi-treated offspring gonads by DIC microscopy and nuclear staining. Control (V) RNAi-treated worms displayed normal gonad morphology (Figure 4b). The normal, linear progression of germ cells from mitosis through meiosis in the C. elegans gonad can also be characterized by nuclear morphology. Normal oogenesis was confirmed by DAPI-staining of control RNAi worms (Figure 4c). Densely packed mitotic nuclei entered meiosis and appeared to organize into an evenly spaced, linear succession during pachytene stages in the distal arm (Figure 4c). As they enter the proximal arm, oocyte nuclei condensed into six bivalent chromosomes during diakinesis, leading to fully grown oocytes. In contrast, ifg-1(RNAi) induced germline deterioration that occurred beyond the reflex (bend) in the gonad leading into the proximal arm. Worms depleted of p170 mRNA (N2) showed relatively normal somatic body size and morphology, but produced few late stage oocytes and no viable embryos. While mitotic and early meiotic nuclei appeared normal, the lack of condensed chromosomes following the diplotene stage suggested that oocytes did not complete diakinesis (Figure 4b and c). N1 (RNAi), which targets both ifg-1 p170 and p130 mRNAs, not only prevented germ cell development but also stunted the somatic growth of the worms. The extent of nuclear development of germ cells was similar to N2 (RNAi). Again germ cells progressed normally through pachytene stages but did not complete diakinesis. Owing to the lack of matured oocytes, no embryos were observed in N2 or C2 (RNAi) worms, nor was there evidence of unfertilized oocytes in the uterus. C2 (RNAi) resulted in two populations of worms. In one population, body size and gonad development was consistent with the larval L2 stage. The other population contained an extended U-shaped gonad consistent with an adult germline despite the small L3-like body size. These results suggest that IFG-1 p170 is required for late meiotic events in the development of oocytes. Because RNAi does not allow us to deplete just the short ifg-1 mRNA, a role for p130 in oogenesis cannot be ruled out.
Induction of germline apoptosis
In wild-type worms, a significant proportion of developing oocytes undergo spontaneous apoptosis. However, these cells are rapidly cleared such that only a small number of apoptotic corpses are apparent in each gonad arm at any time.16 To test whether the observed gonad deterioration involved germline apoptosis, we employed a worm strain expressing a GFP reporter fused to CED-1. CED-1 is a transmembrane receptor on the surface of engulfing cells that recognize apoptotic cell corpses.25 Oocytes that undergo apoptosis become surrounded (decorated) by GFP-expressing gonadal sheath cells. GFP-decorated oocyte corpses were scored in the gonads of RNAi-treated worms. In IFG-1-depleted worms, there were five to eightfold more apoptotic corpses per lobe of the gonad compared to V (RNAi)-treated worms (Figure 5a). Dying germ cells appeared in clusters as several button-like corpses and again coincided with the previously observed proximal area of localized deterioration. Cells undergoing apoptosis also displayed altered nuclear morphology. Due to IFG-1 loss, apoptotic corpses displayed chromatin condensation and marginalization characteristic of cells committed to die (Figure 5a, inset). Increased germ cell death was again observed in late pachytene stages of meiosis I near the bend of the gonadal loop extending to diakinesis. As before, the proximal gonad was devoid of oocyte nuclei indicating a lack of maturing oocytes. These results show that depletion of IFG-1 greatly increases germline apoptotic events and indicate a role for p170 in germ cell maintenance.
Depletion of IFG-1 p170 and p130 causes increased germline apoptosis and induces CED-4 synthesis. (a) DIC and fluorescence images of ced-1∷gfp adult hermaphrodites treated with ifg-1 N2, C2 or control, V (RNAi). White arrowheads indicate clusters of button-like corpses visible by DIC microscopy and in the overlapping GFP fluorescence images (middle row). Note the increased number of corpses in N2 and C2 (RNAi) adults versus control V (RNAi) adults. Bar graph shows a quantitative comparison of germline apoptosis based on the number of GFP-decorated corpses per lobe of the gonad. Error bars display S.E.M. *P-value <0.001; ‘n’ refers to number of lobes included in the count. In C2 (RNAi) worms, clusters of apoptotic cells were so numerous that they blocked entry into the spermatheca. ifg-1(RNAi) ced-1∷gfp worms were co-stained for nuclear morphology with DAPI to show oocyte corpses in which chromatin appeared to marginalize to the inner membrane (inset). This morphology was always coincident with GFP decoration. The number of worms observed by microscopy were: V (n=17), N2 (n=32), and C2 (n=24). (b) Immunostaining of CED-4 expression in the germline of ifg-1(RNAi)-treated worms. Dissected gonads (V’, V, N2 and N1) or decapitated worms (where the gonad was very small; C2, C3) were fixed and stained by immunofluorescence using an anti-CED-4 antibody, and simultaneously for nuclei with DAPI. White arrowheads indicate individual oocytes that contain abundant CED-4 staining. High-magnification images showing the reflex portion of the gonad were obtained using two-channel fluorescence microscopy and merged images are shown (V, N2, N1, C2, C3). Separate images for CED-4 immunostaining and DAPI are shown at low magnification to depict the absence of CED-4 from the entire gonad of vector-treated control worms (V’). The number of worms observed by microscopy were: V (n=3), N2 (n=5), N1 (n=4), C2 (n=4) and C3 (n=7)
Synthesis of pro-apoptotic Apaf-1/CED-4
The pro-apoptotic protein CED-4 is required for both somatic and germ cell deaths in C. elegans.26 CED-4 is the ortholog of mammalian Apaf-1, a core component in the assembly of the apoptosome that ultimately promotes execution by proteolytically activating caspase-3.27 CED-4 was undetectable in either the proximal or distal arms of control RNAi-treated worm gonads that displayed developing oocytes of all stages (Figure 5b). By contrast, immunostaining of gonads dissected from ifg-1(RNAi) worms revealed that CED-4/Apaf-1 becomes strongly expressed in a number of post-pachytene oocytes. CED-4-expressing oocytes were prevalent in gonads from all four RNAi treatments (N2, N1, C2 and C3). Induction of CED-4 by N2 (RNAi) indicated that loss of p170 IFG-1 alone was sufficient to stimulate CED-4 synthesis. CED-4 accumulated in medium sized, post-mitotic oocytes in the proximal gonad arm. This is consistent with the loss of viable, growing oocytes beyond the reflex in the gonads of IFG-1-depleted worms (cf. Figure 4). Furthermore, CED-4 staining showed a punctate pattern, loosely surrounding nuclei of apoptosing germ cells, perhaps indicating that it assembles into large cytoplasmic complexes (apoptosomes). Similar CED-4 localization has been seen in dying cells during embryogenesis.28 In contrast to CED-1:GFP, which decorated shrunken, button-like corpses, CED-4 was clearly abundant in medium to large oocytes (cf. Figure 5a with b). These results provide the first evidence for the involvement of a translation factor in germline apoptosis through the induction of CED-4 synthesis.
ifg-1 gene deletion causes larval developmental arrest
Even with very efficient depletion of ifg-1 mRNA by RNAi, the complete and immediate loss of IFG-1 protein from treated worms is unlikely. To observe the consequences of absolute loss of all ifg-1 expression, we obtained a C. elegans strain heterozygous for a chromosomal deletion of the ifg-1 gene (ok1211) from the Knockout Consortium (Figure 6). The deletion removes 1939 bp from the center of the ifg-1 gene, including over half of the coding capacity of both long and short mRNA forms, and is presumed to be null. The morphological development of homozygous ifg-1(−/−) and heterozygous ifg-1(−/+) offspring was followed over 96 h. Worms lacking functional ifg-1 gene developed to the larval L1/L2 transition (12 h normal development) and then ceased further growth or development (Figure 6b). Pharynx, gut, muscle and gonad structures appeared normal at the time of arrest. Similar to p170 and p130 depletion by C2 RNAi, a dramatic overall growth retardation was observed (cf. Figures 4 and 6). A single functional ifg-1 gene (−/+), however, was sufficient for completely normal growth, development and fertility. In contrast to ifg-1(RNAi) phenotypes, the somatic growth arrest of ifg-1(−/−) worms was matched by arrest of germline proliferation (insets). The gonad of ifg-1(−/−) worms contained only 6–10 mitotic germ cells, indicating that the primordial germ cells (Z2 and Z3) divided just a few times before arresting (Figure 6c). No evidence of meiotic nuclear transition or sperm/oocyte development was evident. Heterozygote gonads, on the other hand, displayed all of the mitotic, transitioning, and meiotic nuclear structures, as well as mature sperm, which characterize wild-type germline development. This indicates that new expression of IFG-1 is required for germline proliferation and somatic growth beyond the L2 larval stage.
Deletion of the ifg-1 gene prevents germ cell proliferation and larval development. (a) Schematic of ifg-1(ok1211) allele showing the deletion of 1939 bp of the gene that removes over half of the coding sequences for both p170 and p130 and is presumed null. Northern blotting (Figure 1) of heterozygous worms suggested there was no stable expression from this allele. (b) DIC images show the extent of development of homozygous worms (ifg-1 −/−) and heterozygous control worms (ifg-1 −/+) after 96 h incubation at 20°C. Size and morphology was identical to heterozygotes (ifg-1 −/+) and wild type worms (not shown) that have just passed the L1/L2 larval transition at 12 h (lower right panel). At higher magnification it was evident that the extent of gonad outgrowth and germ cell proliferation of IFG-1-deficient worms also arrested at the L2 stage (insets). Development of ifg-1(−/−) worms did not progress beyond this stage even after 1 week. These worms nevertheless remained viable, swimming and feeding for at least 1 week (177 h) before becoming lethargic and dying by 3 weeks (not shown). (c) Gonad development was evaluated by DAPI-staining of wild type (ifg-1 +/+), heterozygous (ifg-1 −/+) and homozygous (ifg-1 −/−) worms after 96 or 12 h of development. The gonads (outlined with white dotted line) of IFG-1-deficient worms contained 6–10 germ cell nuclei, similar to control heterozygous or wild type worms (10–12 germ nuclei) at 12 h. In contrast, the gonads of heterozygous worms (ifg-1 −/+; lower right panel) contained many hundreds of germ cell nuclei in all stages of meiosis including the crescent shape characteristic of transition zone (tr) as well as mature sperm (sp) in the spermatheca. For each genotype at least 60 worms were observed to display the phenotype. The number of worms photographed under microscopy were: ifg-1(+/+) (n=11), ifg-1(−/+) (n=12), ifg-1(−/−) (n=9)
Discussion
Two Isoforms of IFG-1 differ in affinity for mRNA caps
The existence of multiple isoforms of mammalian eIF4GI has added to the known heterogeneity (eIF4GI, eIF4GII, p97) of this scaffold protein in eukaryotic cells. The isoforms differ in the extent of N-terminal sequence expressed from the eIF4GI gene via alternate promoters, alternative splicing, and multiple translation initiation codons.21,29 Only one gene (ifg-1, chromosome II) encodes eIF4G in the nematode C. elegans. However, data presented here show that at least two major isoforms of 170 and 130 kDa are expressed from the ifg-1 gene. Both C. elegans IFG-1 isoforms contain the MIF4G motif that allows association with eIF3, eIF4A, and RNA.4,23 p170 contains the complete N-terminal domain that binds eIF4E and PABP, and is consequently enriched in the mRNA cap-binding complex. This isoform associates with some or all of the five eIF4E isoforms, providing further potential to catalyze initiation events that are selective for mRNA recruitment. IFG-1 p170 was distributed similarly in tissues throughout development, whereas eIF4E isoforms segregate between somatic and germ cells.8 IFG-1 p130 lacks this N-terminal portion. Consistent with its domain structure and lack of association with caps, p130 may function like human p97 in a cap-independent complex.12 Thus, alternative initiation complexes may provide the means to control mRNA translation mechanisms to change cell fate.
Depletion of ifg-1 causes a defect in oocyte maturation
We were able to preferentially deplete p170 and not p130 by RNAi using upstream ifg-1 targets. Knockdown of ifg-1 p170 alone caused a significant increase in germline apoptotic events. Our data suggest that IFG-1 p170 is required to complete growth and differentiation steps in late oogenesis. Recent studies have described similar consequences for Drosophila germ cell development of depleting a novel eIF4G isoform (eIF4G2) encoded by the ‘Off-schedule’ gene.17,18 Flies lacking eIF4G2 are defective in spermatogenesis and show a lack of meiotic progression of spermatocytes. We suggest that C. elegans IFG-1 isoforms give rise to alternative translation initiation complexes (eIF4F) that will either possess (p170) or lack (p130) the ability to bind mRNA caps (Figure 7). The relative abundance or activity of these complexes in the C. elegans germline might have consequences similar to the effect of cleaving eIF4G with viral proteases in vivo. Studies in quiescent human cells and Xenopus oocytes have shown that as much as 70% of protein synthesis can be supported by cleaved eIF4G, but cell cycle progression and proliferation are prevented.30,31,32 Forms of eIF4G that contain the extended N-terminus are also much less stable, whereas the C-terminal portion of eIF4G persists for hours following cleavage and sustains cap-independent translation.30 By analogy, the IFG-1 p130 isoform may sustain basic somatic functions in C. elegans, but not promote oocyte development. We postulate that two modes of translation initiation are required to maintain homeostasis in oogenesis (Figure 7).
Model of the translation initiation complexes assembled by eIF4G isoforms p170 and p130 to facilitate alternative translation mechanisms during gametogenesis. (a) During the recruitment of eukaryotic mRNAs to ribosomes for protein synthesis, the p170 eIF4G isoform is able to bind eIF4E (IFEs). The long eIF4G complex catalyzes initiation events necessary for translation of mRNAs required to complete late meiotic development in oocytes. (b) Complexes assembled by the N-terminally truncated p130 are unable to bind IFEs, resulting in the recruitment of mRNAs that can sustain mitotic and early meiotic (mostly nuclear) germ cell development but not completion of the meiotic cell cycle or final phase of oocyte growth. A persistent imbalance in IFG-1 complexes may cause germ cells to enter apoptosis rather than finish oogenesis
IFG-1 p170 is required to suppress germline apoptosis
A few oocytes are naturally undergoing apoptosis at any one time in the adult C. elegans germline.16 Depletion of IFG-1 p170 greatly increased the rate of apoptotic events during pachytene stages resulting in extensive germline degeneration. In the C. elegans germline, programmed cell death is mediated by the anti-apoptotic CED-9/Bcl-2, the Apaf-1 homolog CED-4, caspase-3 homolog CED-3 and engulfment receptor CED-1.25,33 CED-4 acts upstream of caspase activation in the decision-making process.16 Here, we demonstrate that CED-4 expression is induced as a consequence of depleting cap-binding translation initiation complexes. CED-4 protein localized to punctate structures loosely surrounding the nuclei of apoptosing oocytes. CED-4 was previously shown to associate directly with the nuclear envelope in embryonic cells stressed to undergo apoptosis.28,34 The appearance of cytoplasmic CED-4 granules in both cases suggests that apoptosomes assemble in germ cells and embryonic cells that are destined to die. By analogy to Apaf-1 function in mammalian somatic cells, assembly of the apoptosome in oocytes signals the broader activation of executioner proteases such as caspase-3 (CED-3) to drive the completion of apoptosis. Consistent with this hypothesis, the temporal expression of CED-4 in medium to large oocytes occurs prior to changes in nuclear morphology, their shrinkage to ‘button-like’ cells, or their engulfment via CED-1. Therefore, CED-4 is an earlier marker of germ cell apoptosis. Synthesis of CED-4 from maternal mRNA may be linked to changes in IFG-1 isoform function. In mammalian cells apoptosis results in cleavage of eIF4G and p97 by caspase-3 to produce central domain fragments capable of supporting translation of death-associated factors such as p97/DAP5, XIAP, and Apaf-1 via IRES elements in their 5′ UTRs.13 On the basis of these findings, it is possible that IFG-1 isoforms maintain a balance between germline proliferation and apoptosis. Depleting p170 results in increased apoptosis, conceivably due to an increase in the ratio of non-cap-associated p130 complexes to cap-associated p170 complexes (Figure 7). Interestingly, depletion of both mRNAs similarly induced germline apoptosis. By analogy to the stability of eIF4G C-terminus (100 kDa) in Xenopus oocytes,30 it seems reasonable to suggest that p170 protein has a shorter half-life and is outlived by p130, again disrupting the balance between the complexes. Interestingly, preliminary observations indicate that loss of a germline-expressed eIF4E isoform also increases germline apoptosis (MA Richardson and BD Keiper, unpublished observation), further supporting the role of cap recognition in preventing apoptosis.
These data suggest that an isoform shift may alter the balance of translation initiation mechanisms to favor synthesis of pro-apoptotic factors like Apaf-1/CED-4. While no IRES element has been observed in ced-4 mRNA, it is clear that CED-4 expression is induced by ifg-1(RNAi). CED-4 is generally not detectable in the germline, so that CED-4 accumulation in apoptosing oocytes must arise from de novo protein synthesis. In naturally occurring germline apoptotic events, the initial entry into apoptosis might create a positive feedback loop in which p130 either reinforces its own synthesis, as does mammalian p97, or perhaps p130 and p170 become cleaved by CED-3. Cleaved IFG-1 proteins may subsequently reinforce the apoptotic pathway by inducing the synthesis of apoptotic factors such as CED-4 to ensure completion of programmed cell death.
It has been suggested that cytoplasmic rather than nuclear differentiation influences growth and progression of oocytes during meiosis in all animals.35 This suggests a possible role for eIF4G in germline maintenance that links its key position in regulating protein synthesis and the balance of nuclear-to-cytoplasmic contents. The latter has also been suggested as a reason for the natural apoptotic events in the C. elegans germline.16 Dying oocytes are believed to act as nurse cells, providing nutrients and gene products to the sibling oocytes that escape cell death. In the absence of p170, young oocytes are unable to progress beyond diakinesis and complete closure of the cell membrane. The loss of protein synthetic activity may reduce the supply of new cytoplasmic content (proteins) relative to available germ cell nuclei in the ‘measure’ of syncytial contents required to package a mature oocyte. Further germ nuclei deaths may be induced to reestablish balance and rescue sibling germ cells fated to become mature oocytes. A related germline-deficient phenotype was observed in the iffb-1 mutant that lacks eIF5B.19 Together these findings suggest that translation initiation factors exercise primary control over oocyte meiotic progression.
Isoform use in early development and modes of translation initiation
Other developmental roles of IFG-1 were apparent from worms genetically lacking the ifg-1 gene. Curiously, early embryonic lethality was not observed in ifg-1(−/−) offspring. This is likely due to the maternal complement of IFG-1 protein and mRNA synthesized by half of the germ cell population that is ifg-1(+) in the heterozygous ifg-1(−/+) mothers. This ‘half dose’ of IFG-1 is distributed evenly to all developing oocytes in the syncytial gonad. Pre-existing protein as well as maternal ifg-1 mRNA are presumably sufficient for p130 and p170 functions throughout embryogenesis. However, growth beyond the larval L2 stage requires additional IFG-1 translational activity for somatic growth (body size) and gonad development (germ cell differentiation). Our observations suggest two developmental stages with a critical requirement for IFG-1: late meiotic oocyte development and L2 larval growth. Not surprisingly, these developmental stages are known for translational control events and enhanced protein synthesis. Such observations are no longer consistent with the classical dogma that the translation initiation apparatus is merely part of the cellular housekeeping machinery.
Materials and Methods
Strains
C. elegans strain N2 var. Bristol was used as wild type and were cultured as described by Brenner, 1974.36 Growth was at 20 °C on NGM plates with E. coli strain OP50 or strain HT115 bearing plasmids to direct dsRNA synthesis unless otherwise noted.36 The null mutant allele ifg-1(ok1211) originally identified by the Vancouver C. elegans Knockout Consortium was created by chemical mutagenesis and deleted 1939 bp of the ifg-1 gene. The isolate was obtained from the Caenorhabditis Genetics Center and outcrossed six times to wild type to remove any secondary mutations, then crossed into a gfp-expressing chromosome II-balancer strain to generate the stable heterozygous strain KX38 [ifg-1(ok1211)/mIn1 [mIs14 dpy-10 (e128)] II]. Single worm genomic PCR (Forward 5′-ACCCTTGTGACGCCTGTTTC-3′, Reverse 5′-TCACCAGTAACGGTCCACAA-3′) was used to monitor the ifg-1 deletion and wild type alleles. ifg-1(ok1211) homozygous worms were obtained by self-crossing KX38 progeny carrying the deletion and picking embryos and larvae lacking GFP fluorescence to fresh seeded NGM plates for observation at 20°C. Other strains analyzed in subsequent experiments include MD701, (bcIs39V [Plim-7 ced-1∷gfp and lin15(+)])25 generously provided by Dr Barbara Conradt (Dartmouth University)
Identification of the ifg-1 gene
Blast searches of the C. elegans genome database were conducted with the primary amino-acid sequences of human eIF4GI, eIF4G-II, p97, Drosophila eIF4G-1, yeast TIF46311 and TIF4632, and Arabidopsis eIF4G-1 and eIF4G-2. All searches identified a single C. elegans gene, M110.4 on chromosome II, as encoding the closest eIF4G ortholog. Pairwise alignments displayed about 23% amino-acid sequence similarity (12–14% identity) throughout their entire lengths, including 29–33% identity in the central MIF4G domain. This gene encodes nine putative exons and was subsequently renamed ‘ifg-1’ for initiation factor 4G gene 1 (Figure 1a, WormBase: C. elegans online database. 19 June 2007, cited; release WS176; Available from: http://www.wormbase.org, J Hodgkin personal communication). Unlike yeast and other higher eukaryotes, no second gene encoding eIF4G has been found in the C. elegans genome by various sequence homology searches.
Purification and analysis of RNA from C. elegans
Total RNA was purified from frozen worms ground in liquid nitrogen and solubilized in four volumes of Trizol (Invitrogen) according to the manufacturer's procedure. Further purification of RNA by extraction from Tris/urea/SDS buffer was performed as previously described.8 For northern blotting, 20 μg of total RNA was resolved on 1% agarose/formaldehyde gels and blotted to NitroPlus 2000 membrane (MSI Separations) and hybridized to antisense RNA probes as described.30 mRNA sizes were determined using a linear-log plot of migration versus nt length using the known lengths of C. elegans 18S and 28S rRNAs as standards. RNase protection was performed on 20 μg of total RNA using the RPAIII kit (Ambion) according to the manufacturer's directions. Protected fragments were resolved on 6% acrylamide (19 : 1) Tris/borate/EDTA gels. In both RNA assays, the probe was an antisense RNA transcribed from the plasmid pSKifg-deltaSX containing bp 24–2083 of the full length ifg-1 cDNA uniformly labeled with α-[32P]UTP (ICN).
Purification of cap-binding proteins from C. elegans
The mRNA cap-binding complex was purified from snap-frozen worms ground in liquid nitrogen by affinity chromatography on m7GTP-Sepharose (Pharmacia) as previously described.6
IFG-1 antibody production
Rabbit antiserum against the peptide KRQRKALEIVDPTTKKA corresponding to sequence near the N-terminus of IFG-1 (amino acids 136–152) has been described7 and was generously provided by Dr Robert E Rhoads (Louisiana State University HSC). Rabbit antisera to the central domain (amino acids 396–762) of IFG-1 were prepared as follows: plasmid pGST-E6 contained 1102 bp from the middle of full-length ifg-1 cDNA (bp 1209–2310) subcloned into the plasmid pGEX4T-2 (Pharmacia). Recombinant E6-GST fusion protein was prepared from BL21(DE3)pLysS E. coli cells bearing the plasmid pGST-E6. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added to 3 mM to induce the fusion protein production. After centrifugation, sedimented cells were suspended in phosphate-buffered saline Tris (PBST), 1 mM PMSF, 1 mM DTT, Complete® protease inhibitor tablet (Boehringer Mannheim) and sonicated on ice in short 30 s bursts. To the sonicated cells was added glutathione sepharose 4B, 50% slurry/1 × PBST. After sedimentation, bound fusion protein was eluted directly from the column with 10 mM glutathione, 50 mM Tris pH 8.0. Further purification of the E6-GST protein was performed using heparin sepharose and elution in 600 mM KCL. Protein fractions were pooled and concentrated. Two rabbits were immunized and sera provided by Rockland Immunochemicals. Antisera to E6 IFG-1 from two independent rabbits (E6-23 and E6-24) were shown to react similarly to both p170 and p130 from C. elegans lysates. Preimmune sera from both rabbits failed to react with either isoform.
Western blot analysis
Frozen worm pellets were boiled for 5 min at 100°C in 4 × SDS sample buffer, centrifuged at 15 000 × g for 10 min, and supernatant corresponding to 20 μg lysate protein resolved by electrophoresis 8% (30 : 0.8) polyacrylamide SDS-PAGE. Proteins were electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes and incubated with a 1 : 1000 or 1 : 2000 dilution of appropriate primary antibody in TST (10 mM Tris-HCl ph 7.4, 150 mM NaCl, 0.05% Tween 20) containing 5% dry non-fat milk. Blots were washed extensively and incubated 1 : 2000 goat anti-rabbit IgG secondary antibody conjugated to horseradish peroxidase. Detection was by ECL+ kit (GE Healthcare/Amersham) using a Typhoon 9410 Fluorescence/Phosphorimaging scanner at the ECU PhIFI Core Facility.
RNA interference by feeding
Plasmids were constructed using standard cloning techniques. The following fragments of the ifg-1 cDNA were subcloned into plasmid pL4440:24 pT72ifgN2 contained 398bp (nt 24–479) of sequence the ifg-1 cDNA; pT72ifgN1 contained 301bp (nt 1115–1508); pT72ifgC2 contained 626bp (nt 2080–2705); and pT72ifgC3 contained 371bp (nt 2706–3091). ifg-1 double-stranded RNA (dsRNA) for each of the targets was expressed in E. coli strain HT115(DE3) in cultures containing ampicillin and tetracycline and plated on standard NGM/agar media supplemented with IPTG and ampicillin. Wild type or CED-1∷GFP bearing worms were synchronized by washing off plates with 1.2% hypochlorite 0.5 M NaOH solution, rinsing in M9, and transferring pellets to OP50 seeded plates. Worms were incubated at 20°C until L3/L4 stages just prior to formation of a pronounced gonad. Five to six P0 hermaphrodites were transferred to seeded RNAi plates containing bacteria expressing appropriate target dsRNA or control pL4440 vector dsRNA (187 bp). Adults were allowed to feed and lay offspring (F1 s) for 24–36 h at 25°C before being transferred to subsequent plates. Alternatively, RNAi-feeding treatments were performed as described by Kamath et al.37 Briefly, a single colony of HT115 containing the appropriate cloned vectors was grown in 2 × YT media containing 100 μg/ml ampicillin (Amp) overnight at 37°C. Small aliquots were diluted 1 : 10 in 2 ml cultures and grown for 8 h, then seeded onto NGM plates containing 1 mM IPTG and 100 μg/ml Amp. Plates were allowed to dry and were incubated at 22°C overnight. Three plates were assayed for each target. F1 phenotypes were scored over a 72 h period.
Microscopy
ifg-1(RNAi) animals were immobilized in 20 mM sodium azide in buffer M9 and examined using a Zeiss Axiovert 200 M Differential Interference Contrast (DIC) microscope equipped with an Axiocam MRM2 CCD camera and corresponding DAPI/Hoechst, and FITC/GFP optics and F-fluar 40 × and Plan Neofluar 100 × objectives. For nuclear staining, worms were fixed in MRWB (80 mM KCl, 20 mM NaCl, 10 mM disodium EGTA, 5 mM spermidine-HCl, 15 mM PIPES pH 7.4, 25% methanol, 1.6% formaldehyde) and placed at −80°C for at least 20 min. Samples were thawed on ice and worms pelleted briefly by centrifugation at 1000 r.p.m. in a microcentrifuge. Supernatant was replaced by 1 mg/ml DAPI in M9. Images were collected and analyzed using Axiovision LE 4.3 software. Scaling was performed by calibration of software measurements with a 0.010 mm stage micrometer (Klarmann Rulings Inc.). Where noted, worms were incubated in 20 mM sodium azide, 5 μg/ml Hoechst 33342 in M9 for 1 h at 20°C prior to microscopy.
CED-4 Immunostaining
CED-4 localization in the germline was determined with immunostaining using the anti-CED-4 (cN-21) goat polyclonal antibody (Santa Cruz Biotechnology Inc.) diluted 1 : 50. RNAi-treated worms were washed 0.75 × M9 with 10 mM levamisole (MP Biomedicals). Worm heads were removed by cutting just below the pharynx, and where possible, the gonad was teased out of the carcass. Gonad and carcass were transferred to a poly-L-lysine-treated slide with cover slip and exposed to dry ice for 10 min. After removal of the cover slip the slide was submerged in methanol at −20°C for 10 min and acetone at −20°C for 10 min. Following three washes in TST solution, primary antibody was applied overnight at 4°C. The slides were washed in TST and the secondary antibody, donkey anti-goat IgG Alexa 488 (Molecular Probes) diluted 1 : 1000 was applied 1–2 h at room temperature. The samples were washed in a TST-containing DAPI and mounted in a 90% glycerol/DABCO (Arcos) media. Microscopy and imaging were performed as mentioned above.
Micro-scale RNA isolation, RT-PCR and qRT-PCR
Fifty late larval/adult F1 worms were collected into 10 μl of single worm lysis buffer (15 mM Tris pH 8.0, 50 mM KCl, 1 mM MgCl2, 0.45% NP-40, 0.45% Tween 20) containing 0.1 mg/ml proteinase K (60 min, 60°C; 10 min, 95°C). Upon completion of lysis, RNA was then isolated using the RNeasy Mini-Kit (Qiagen) with 20 μg glycogen carrier, treated with DNase I, and resuspended in 15–50 μl of DEPC-treated water for RT-PCR or qRT-PCR.
Quantitative RT-PCR was conducted using Invitrogen One Step Superscript III Platinum/Taq HiFi and standard polymerases. The optimal annealing temperature (Ta), number of cycles, and RNA dose response were determined in multiple preliminary assays to generate an amplification signal within a linear range of response for both ifg-1 amplicons and the act-1 amplicon (data not shown). Optimal Ta and number of cycles were determined by testing equivalent worm concentrations across five different cycles and five different temperatures, respectively. It was determined using a temperature gradient that the following Ta conditions were optimal for each set of ifg-1 primers: 53°C for upstream; and 55°C for downstream. The optimal number of cycles was determined by a similar method and shown to be 40 cycles for each primer set. Next, the optimal input sample RNA amount was determined using fivefold serial dilutions of worm RNA equivalents 0.004, 0.02, 0.1, 0.5 and 2.5 using the optimal Ta for each primer set and appropriate number of cycles. Total amount of RNA equivalent to one worm was adequate in measuring expression of ifg-1. Furthermore, there was an obvious linear relationship of ifg-1 mRNA under these conditions. Final conditions were as follows: amplification using total RNA equivalent to one worm was sufficient for ifg-1 detection followed by a Ta=55°C and 40 cycles. Similar optimization was done using primers complementary to an internal control mRNA, act-1. Optimization revealed a Ta=60 °C and 40 cycles to be optimal for act-1 amplification; however, only 0.1 worm equivalents were necessary to achieve similar amplicon signals. Actin mRNA, like other housekeeping genes, has been shown to be present at very high levels and as such reaches a plateau faster than less abundant RNAs.38 Primer sets (MWG Biotech Inc.) used for analysis of ifg-1 mRNA for amplication were upstream (5′-AATGAACCAGTCGGCGTATC-3′ and 5′-CAACTTGTGGAGCTATGGATGC-3′) and downstream (5′-ACCAAACTGGGCAAACAAAG-3′and 5′-GCGCATCGTTGGAGGATACA-3′) corresponding to upstream and downstream portions of the full length mRNA. Actin primers (5′-TGACGACGAGGTTGCCGCTCTT-3′ and 5′-GGACTGGGTGCTCTTCTGGG-3′) were used as an endogenous mRNA control.
For real time PCR (qRT-PCR), reverse transcripts were created using 20–100 ng of total RNA and the iScript cDNA Synthesis Kit (Biorad). Quantification of transcripts was performed using iCycler iQ Real-Time PCR with iQ SYBR Green supermix (Biorad). Primer sequences include: ifg-1 forward 5′-TCAGACACAACCACCACTAC-3′ and reverse 5′-TCCTCTTTTGATTCTTCCTTT-3′; gpd-3 (GAPDH) forward 5′-TCCAGTACGATTCCACTCAC-3′ and reverse 5′-GATCTCAGCTGGGTCTCTT-3′. Reactions were conducted in triplicate from two independent preparations of RNA, quantified and normalized to gpd-3 (endogenous control) using the 2−ΔΔCt method.
Abbreviations
- eIF:
-
eukaryotic translation initiation factor
- ifg-1 :
-
initiation factor 4G gene 1
- p97/DAP5:
-
human 97 kDa eIF4G-homologous protein, death-associated protein 5
- Apaf-1/CED-4:
-
apoptotic peptidase-activating factor 1/cell death abnormality gene 4 protein
- 4EHP:
-
eIF4E-homologous protein
- IRES:
-
internal ribosome entry site
- m7GTP:
-
7-methylguanosine triphosphate
- RT-/qRT-PCR:
-
reverse transcriptase- or real time polymerase chain reaction
- RNAi:
-
RNA interference
- dsRNA:
-
double-stranded RNA
- PABP:
-
poly(A)-binding protein
References
Dworkin MB, Dworkin-Rastl E . Functions of maternal mRNA in early development. Mol Reprod Dev 1990; 26: 261–297.
Mendez R, Richter JD . Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001; 2: 521–529.
Meric F, Hunt KK . Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther 2002; 1: 971–979.
Keiper BD, Gan W, Rhoads RE . Protein synthesis initiation factor 4G. Int J Biochem Cell Biol 1999; 31: 37–41.
Hernandez G, Altmann M, Sierra JM, Urlaub H, del Corral RD, Schwartz P et al. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech Dev 2005; 122: 529–543.
Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE et al. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 2001; 128: 3899–3912.
Dinkova TD, Keiper BD, Korneeva NL, Aamodt EJ, Rhoads RE . Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol Cell Biol 2005; 25: 100–113.
Keiper BD, Lamphear BJ, Deshpande AM, Jankowska-Anyszka M, Aamodt EJ, Blumenthal T et al. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J Biol Chem 2000; 275: 10590–10596.
Miyoshi H, Dwyer DS, Keiper BD, Jankowska-Anyszka M, Darzynkiewicz E, Rhoads RE . Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J 2002; 21: 4680–4690.
Syntichaki P, Troulinaki K, Tavernarakis N . eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 2007; 445: 922–926.
Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S et al. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18: 334–342.
Hundsdoerfer P, Thoma C, Hentze MW . Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci USA 2005; 102: 13421–13426.
Nevins TA, Harder ZM, Korneluk RG, Holcik M . Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J Biol Chem 2003; 278: 3572–3579.
Marissen WE, Lloyd RE . Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol 1998; 18: 7565–7574.
Metzstein MM, Stanfield GM, Horvitz HR . Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet 1998; 14: 410–416.
Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO . Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 1999; 126: 1011–1022.
Baker CC, Fuller MT . Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 2007; 134: 2863–2869.
Franklin-Dumont TM, Chatterjee C, Wasserman SA, Dinardo S . A novel eIF4G homolog, Off-schedule, couples translational control to meiosis and differentiation in Drosophila spermatocytes. Development 2007; 134: 2851–2861.
Yu X, Vought VE, Conradt B, Maine EM . Eukaryotic translation initiation factor 5B activity regulates larval growth rate and germline development in Caenorhabditis elegans. Genesis 2006; 44: 412–418.
Ciosk R, DePalma M, Priess JR . Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 2006; 311: 851–853.
Bradley CA, Padovan JC, Thompson TL, Benoit CA, Chait BT, Rhoads RE . Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J Biol Chem 2002; 277: 12559–12571.
Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ et al. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 2007; 6: 111–119.
Korneeva NL, Lamphear BJ, Hennigan FL, Merrick WC, Rhoads RE . Characterization of the two eIF4A-binding sites on human eIF4G-1. J Biol Chem 2001; 276: 2872–2879.
Timmons L, Court DL, Fire A . Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 2001; 263: 103–112.
Zhou Z, Hartwieg E, Horvitz HR . CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 2001; 104: 43–56.
Hengartner MO, Horvitz HR . Programmed cell death in Caenorhabditis elegans. Curr Opin Genet Dev 1994; 4: 581–586.
Vaux DL . CED-4 – the third horseman of apoptosis. Cell 1997; 90: 389–390.
Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y et al. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 2000; 287: 1485–1489.
Han B, Zhang JT . Regulation of gene expression by internal ribosome entry sites or cryptic promoters: the eIF4G story. Mol Cell Biol 2002; 22: 7372–7384.
Keiper BD, Rhoads RE . Cap-independent translation initiation in Xenopus oocytes. Nucleic Acids Res 1997; 25: 395–403.
Keiper BD, Rhoads RE . Translational recruitment of Xenopus maternal mRNAs in response to poly(A) elongation requires initiation factor eIF4G-1. Dev Biol 1999; 206: 1–14.
Perez L, Carrasco L . Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology 1992; 189: 178–186.
Zornig M, Hueber A, Baum W, Evan G . Apoptosis regulators and their role in tumorigenesis. Biochim Biophys Acta 2001; 1551: F1–37.
Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y . Matefin/SUN-1 is a nuclear envelope receptor for CED-4 during Caenorhabditis elegans apoptosis. Proc Natl Acad Sci USA 2006; 103: 13397–13402.
Fulka Jr J, First NL, Moor RM . Nuclear and cytoplasmic determinants involved in the regulation of mammalian oocyte maturation. Mol Hum Reprod 1998; 4: 41–49.
Brenner S . The genetics of Caenorhabditis elegans. Genetics 1974; 77: 71–94.
Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J . Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2001; 2: Research 0002.
Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G . Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online 2001; 3: 19–25.
Acknowledgements
We thank Dr Yugi Kohara (NIG, Japan) for several ifg-1 cDNAs, the C. elegans Knockout Consortium and the Caenorhabditis Stock Center for the initial ifg-1(ok1211) strain. In addition, we thank Dr Barbara Conradt (Dartmouth University) for the ced-1∷gfp strain, Dr Andrew Fire (Stanford University) for plasmid pL4440 and the HT115 bacterial strain, Dr Jennifer Schisa (University of Central Michigan) and Dr Jennifer Tenlan (University of North Carolina, Chapel Hill) for advice on immunostaining, and Dr Robert E Rhoads (Louisiana State University HSC) for the anti-N-terminal IFG-1 antibody. This work was supported by Grants IRG 5-89812 from the American Cancer Society and MCB-0321017 from the National Science Foundation to BDK.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Piacentini
Rights and permissions
About this article
Cite this article
Contreras, V., Richardson, M., Hao, E. et al. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ 15, 1232–1242 (2008). https://doi.org/10.1038/cdd.2008.46
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.46
Keywords
This article is cited by
-
Molecular targets for modulating the protein translation vital to proteostasis and neuron degeneration in Parkinson’s disease
Translational Neurodegeneration (2019)
-
Phasing in on the cell cycle
Cell Division (2018)
-
Heterogeneity and specialized functions of translation machinery: from genes to organisms
Nature Reviews Genetics (2018)