Abstract
Histone acetylation induces chromatin opening by perturbing higher-order chromatin compaction and folding, suggesting that histone acetylation and deacetylation dynamics are central to chromosome condensation or decondensation. The condensation of chromosomes during mitosis is an essential prerequisite for successful chromosome segregation. In this study, we depleted three representative histone acetyltransferases (HATs; p300, CBP, and P/CAF) using shRNAs to explore their role in regulating mitotic progression and chromosome segregation. We showed that HAT depletion severely interfered with the normal timing of mitotic progression, and it reduced condensin subunit levels. The predominant response to HAT depletion, in both human primary and cancer cells, was a mitotic catastrophe following aberrant mitotic arrest. Alternatively, adaptation to HAT depletion, particularly in cancer cells, led to multinucleation and aneuploidy. Interestingly, mitotic catastrophe induced by HAT depletion appeared to be coupled to the signaling process of H2AX phosphorylation and foci formation, independently of DNA double-strand breaks and DNA damage. Taken together, our results provide novel molecular evidence that HAT proteins maintain mitotic chromatin assembly and integrity as a cellular determinant of mitotic cell death.
Similar content being viewed by others
Main
Histone modifications involving reversible acetylation and deacetylation events play an important role in transcription, DNA replication, and chromosome segregation.1, 2 Acetylation occurs on conserved lysine residues of core histones, and it induces chromatin opening by perturbing higher-order chromatin compaction and folding. Chromosome condensation occurs during the G2 phase of the cell cycle, with compaction becoming apparent during prophase and reaching a maximum during metaphase. Chromosome decondensation begins in the following stage, anaphase. These processes are essential to correctly separate parental genetic information into the two daughter cells during subsequent nuclear division. It is generally accepted that this dynamic equilibrium between chromosome condensation and decondensation is directly related to the degree of histone deacetylation and acetylation, which are, in turn, governed by the activities of histone deacetylases (HDACs) and histone acetyltransferases (HATs), respectively.2
Despite evidence linking HATs with transcriptional regulation, the exact role of HATs in maintaining an active chromatin configuration during the transcription-silencing mitotic stage of the cell cycle is not completely understood. Recent studies have established that HDAC activity plays an important role in regulating mitotic checkpoint activation, kinetochore assembly, and cytokinesis.3, 4, 5 Consequently, aberrant control of HDAC activity contributes to defects in the mitotic spindle checkpoint, failure of appropriate chromosome segregation, and chromosome instability. However, previous studies have relied heavily on HDAC inhibitors, such as trichostatin A and SAHA, to examine the effects of histone acetylation or deacetylation on mitotic chromatin remodeling.4, 5, 6 Although HDAC inhibitors produce the expected histone hyperacetylation state, they exert additional effects that complicate the interpretation of changes in chromatin status. Through transcriptional effects and post-translational modifications, these agents can alter the expression of genes or the activity of proteins involved in the mitotic cell cycle progression, rather than directly regulating mitotic chromatin condensation or decondensation.
Mitosis is a dramatic event in the life of a cell, irreversibly distributing duplicated chromosome pairs into the daughter cells. As such, it is no surprise that exquisitely fine-tuned regulatory mechanisms have evolved to ensure that each step in the process is completed before the next begins. The mitotic checkpoint helps to maintain the fidelity of this process, ensuring accurate chromosome segregation by sensing that the kinetochores of sister chromatids have successfully attached to spindle microtubules and are under adequate tension.7, 8 In normal cells, the operation of this checkpoint activates signaling cascades that delay anaphase onset until these criteria are satisfied, thereby preventing the chromosome instability and polyploidy that are hallmarks of cancer.9, 10 Mitotic catastrophe is a cell death mode occurring either during or shortly after a dysregulated/failed mitosis and can be accompanied by morphological alterations, including micronucleation and multinucleation (reviewed by Kroemer et al.11). Recently, mitotic catastrophe has been widely reported as an effect of conventional chemotherapeutic drugs, such as the antimicrotubule agents taxans and vinca alkaloids, and by IR- and DNA-damaging drugs.12 Although the term ‘mitotic catastrophe’ has come to describe a new form of cell death that results from abnormal mitosis, there is still no broadly accepted definition.
As a first step toward understanding the role of HAT proteins in mitotic progression and chromosome segregation, we used RNA interference to deplete three representative HATs (CBP, p300, and P/CAF) in human cancer cells and noncancerous primary adult stem cells. Our results clearly indicated that the predominant response to HAT depletion is mitotic catastrophe followed by H2AX phosphorylation and the reduction of condensin expression.
Results
Depletion of HAT severely interfered with the normal timing of mitotic progression
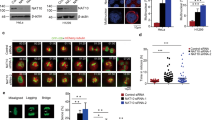
To explore the role that HAT plays in the regulation of mitotic progression and chromosome segregation, we used RNA interference to deplete HeLa cells of three representative HATs (p300, CBP, and P/CAF). To accomplish this, we transfected HeLa cells with plasmids encoding shRNA against p300, CBP, or P/CAF, which resulted in a reduction of the original cellular levels of these HATs by approximately 70% 48 h after transfection (Figure 1a). To visualize the chromosomes, we cotransfected HeLa cells with green fluorescence protein (GFP)-tagged H2B (GFP-H2B) (Figure 1b, upper panel). We considered the point at which the nuclear envelope broke down, which occurred as each cell entered mitosis, as t0. Next, we determined the time from t0 required for individual cells to complete chromosome separation. In cells transfected with the empty control plasmid, the average time required to complete chromosome separation was 63.5±9.8 min (Figure 1b, lower panel). In contrast, HeLa cells transfected with p300 shRNA, CBP shRNA, or P/CAF shRNA exhibited significant delays in chromosome separation, requiring an average of 78.2±21.5, 75.2±18.4, and 74.2±18.4 min for separation to occur, respectively (Figure 1b, lower panel). Interestingly, these HAT-depleted cell populations also exhibited increased frequencies of cells with aberrantly delayed and accelerated mitotic cell cycle durations, which is typical of cells with mitotic defects. Specifically, approximately 43, 33, and 45% of the p300-, CBP-, and P/CAF-depleted cells exhibited aberrant mitotic durations, respectively, whereas the duration of mitosis was abnormal in only approximately 3% of the control cells (Figure 1c). In addition, separate experiments conducted using two different shRNAs to simultaneously deplete pairs of HATs in HeLa and HCT116 cells yielded very similar results (data not shown). Furthermore, flow cytometric analyses using asynchronized HeLa cells in which p300, CBP, or P/CAF had been depleted showed a significant increase in the proportion of G2/M cells and a reduced number of G1 cells when compared with the control cells (Supplementary Figure S1).
Depletion of HAT interfered with the normal timing of mitotic progression and sustained mitotic arrest. (a) HeLa cells were transiently transfected with control luciferase shRNA or two independent shRNAs targeting p300 (p300 shRNAs # 1 and # 2), CBP (CBP shRNAs # 1 and # 2), or P/CAF (P/CAF shRNAs # 1 and # 2), as described in the Materials and Methods. Extracts of the transfected cells were then separated by SDS-PAGE and immunoblotted with anti-p300, anti-CBP, anti-P/CAF, and anti-actin. (b) HeLa cells were transfected with a plasmid encoding H2B-GFP and control luciferase shRNA, p300 shRNA, CBP shRNA, or P/CAF shRNA. Cells were then cultured and the mitotic progression was imaged by time-lapse microscopy. The times from nuclear envelope breakdown to complete chromosome separation are indicated. Representative time-lapse images of control shRNA-transfected cells are shown (upper panel). Mitotic progression data from randomly selected cells are presented (lower panel). (c) The estimated frequency of cells with aberrant mitotic durations (<60 min or approximately >95 min) was based on counts of approximately 220 mitotic phase cells from each shRNA transfection obtained from three independent experiments. (d) HeLa cells were transfected with control pSuper (pS) or pSuper-p300 shRNA (pS-p300 shRNA), -CBP shRNA (pS-CBP shRNA), or -P/CAF shRNA (pS-P/CAF shRNA), after which the cells were synchronized by nocodazole treatment (N; 100 ng/ml) or nocodazole treatment and released into the cell cycle (NR). The cells were then harvested at the indicated times and then immunoblotted with anti-Mad2, anti-phospho H3, anti-securin, anti-BubR1, and anti-Bub3
Next, we compared the expression profiles of the mitotic marker proteins, phosphorylated histone H3 (phospho-H3), securin, and BubR1, in control and HAT-depleted cells. As earlier, we transfected HeLa cells with control shRNA, p300 shRNA, CBP shRNA, or P/CAF shRNA and then synchronized the cells with nocodazole treatment (N) or nocodazole treatment (NR)/post-release. At the indicated times, we harvested the cells and then immunoblotted them with antibodies against Mad2, phospho-H3, securin, BubR1, or Bub3 (Figure 1d). Interestingly, the control cells exhibited markedly reduced levels of phospho-H3 and securin at 2.5 h NR, whereas the corresponding levels of these two mitotic marker proteins in p300-depleted cells did not differ significantly throughout the experimental period. Similar results were observed when CBP- and P/CAF-depleted cells were evaluated (data not shown). Furthermore, the levels of two mitotic checkpoint proteins, BubR1 and Mad2, were stable in p300-depleted cells when compared with the levels in the control cells (Figure 1d). In addition, we evaluated the subcellular localization of the mitotic checkpoint proteins, Bub1, BubR1, and Mad2, in HAT-depleted cells. As expected, depletion of each HAT protein led to a significant increase in the number of misaligned chromosomes. Moreover, subsequent immunofluorescence analyses also showed the kinetochore localization of Bub1, BubR1, and Mad2 at misaligned chromosomes, which is the signal to activate the mitotic checkpoint. Finally, these HAT-depleted cells exhibited a significant increase in the population of multipolar spindles (from 5% in the control shRNA-transfected cells to 15–18% in the treated cells) (Figure 2b and c). Taken together, these data strongly suggest that depleting HAT proteins severely impaired the normal timing of mitotic progression following sustained activation of the mitotic checkpoint.
Cells with depleted p300, CBP, or P/CAF levels exhibited severe mitotic defects. (a) HeLa cells were transfected with p300 shRNA, CBP shRNA, P/CAF shRNA, or control luciferase shRNA. Forty-eight hours after transfection, the cells were fixed, permeabilized, and then immunostained with antibody against BubR1, Bub1, or Mad2, and CREST serum. DNA was visualized with Hoechst dye. (b) To analyze the mitotic spindles, HeLa cells were transfected with luciferase (Luc) shRNA, p300 shRNA, CBP shRNA, or P/CAF shRNA. Forty-eight hours post-tansfection, the cells were immunostained with anti-α-tubulin antibody (green) and CREST serum (red). DNA was visualized with Hoechst dye (blue). Representative confocal microscopic images are shown. Cells with more than two poles were considered as multipolar (abnormal mitotic) cells. (c) Graphical comparison of multipolar spindle numbers in cells with depleted levels of p300, CBP, or P/CAF. The values shown are the means of three independent experiments, in which more than 300 mitotic phase cells were counted, and the bars indicate the standard deviations
Depletion of HAT led to sustained mitotic arrest followed by severe mitotic catastrophe
To further examine the morphological phenotype of cells lacking HAT, we cotransfected HeLa cells with an expression plasmid encoding an H2B-GFP fusion protein and shRNA against p300, CBP, or P/CAF. Time-lapse microscopic analyses at the single-cell level revealed that most control shRNA-transfected cells displayed normal chromosome segregation and cell division (Figure 3a). However, a substantial proportion of the HAT-depleted cells were arrested in (pro)metaphase and unable to progress into the cell cycle. As a result, these aberrantly delayed cells ultimately underwent mitotic catastrophe (Figure 3a). A consistently high percentage of cells exhibited features of mitotic catastrophe following transient depletion of HAT (approximately 34, 32, or 25% of the total HeLa cell population following knockdown of p300, CBP, or P/CAF, respectively; Figure 3b, lanes 1–4). Similarly, when 116 colorectal cancer cells were evaluated, approximately 32, 33, and 25% of p300 shRNA-, CBP shRNA-, and P/CAF shRNA-transfected cells, respectively, underwent mitotic catastrophe (Figure 3b, lanes 5–8). Together, these data clearly show that loss of the HATs resulted in a severe aberrant delay in the metaphase-to-anaphase transition that led to progression to mitotic catastrophe.
Depletion of HAT led to mitotic catastrophe following sustained mitotic arrest. (a and b) HeLa and HCT116 cells were cotransfected with an H2B-GFP expression plasmid and an shRNA specifically targeting p300, CBP, P/CAF, or luciferase (control). The cells were cultured and the mitotic progression was imaged by time-lapse microscopy. Representative time-lapse images of cells transfected with each shRNA are shown (a). The frequency of cells (HeLa, left graph; HCT116, right graph) undergoing mitotic catastrophe following depletion of p300, CBP, or P/CAF is shown (b). At least 100 cells/sample were counted per experiment, and the experiment was performed at least three times. (c and d) HeLa cells were cotransfected with an H2B-GFP expression plasmid and shRNA specifically targeting p300, CBP, P/CAF, or luciferase (control). Cells were immunostained with Alexa-conjugated anti-phospho-H3 (P-H3), anti-active caspase 3 (caspase 3a), and Hoechst dye (c). Data represent the average number of cells that were positive for both phospho-H3 and caspase 3a as determined by three independent experiments; 100 cells/sample were counted within each experiment (d)
Recent studies have suggested that mitotic catastrophe is a type of cell death that results from abnormal mitosis, which usually culminates in the formation of large aneuploid cells with multinucleation.13 Mitotic catastrophe may be perceived as a molecular device that prevents aneuploidization following mitotic checkpoint activation. Typical features of irreversible mitotic catastrophe include cell ‘blebbing,’ formation of micronuclei, and chromosome fragmentation. Importantly, the process of mitotic catastrophe appears to be mediated by both caspase-dependent and -independent pathways, indicating that this pathway is distinct from the process of typical apoptotic cell death.
It has been reported that cells undergoing mitotic catastrophe are usually positive for MPM2 and phospho-H3. We found that the population of control cells in mitotic phases defined by antiphospho-H3 and DNA staining were largely devoid of activated caspase 3, as evidenced by the largely negative response to caspase 3a antibody (Figure 3c). However, transient knockdown of p300, CBP, or P/CAF expression induced a significant increase in the frequency of caspase 3a-positive cells. Consequently, only approximately 6.8% of control cells, but 23.2, 21.2, and 22.4% of p300-, CBP-, and P/CAF-depleted cells, respectively, were immunoreactive for activated caspase 3 (Figure 3c and d). Taken together, these data strongly support the idea that loss of HAT activity induced severe mitotic catastrophe.
Adaptation to HAT depletion induced multinucleation and aneuploidy in cancer cells
In addition to severe mitotic catastrophe, a striking feature of HAT-depleted cells was the generation of multinucleated and aneuploid cells. To expand on our initial observations, we stained mitotic chromosomes from control and HAT-depleted HeLa cells with DAPI. As shown in Figure 4a, p300-depleted cells underwent dramatic changes in nuclear morphology, exhibiting multinucleation, large nuclei, and/or multiple micronuclei. Similar results were observed following CBP or P/CAF depletion (data not shown). However, HAT knockdown also increased the susceptibility of the cells to apoptotic cell death and/or mitotic catastrophe during the process of chromosome segregation. In spite of this, approximately 19, 21, and 23% of p300 shRNA-, CBP shRNA-, and P/CAF shRNA-transfected HeLa cells, respectively, successfully adapted to HAT depletion and showed aberrant nuclear morphology, compared with approximately 6% of the vector-transfected control cells (Figure 4b). Similarly, when HCT 116 cells were used, approximately 17, 18, and 16% of p300 shRNA-, CBP shRNA-, and P/CAF shRNA-transfected cells, respectively, exhibited disorganized and abnormal mitotic chromosome morphology (Figure 4c).
HAT depletion resulted in multinucleation and aneuploidy in cancer cells. (a) Panels show control and p300-depleted HeLa cells 2 days after transfection with shRNAs. Nuclei were stained with Hoechst dye. (b) Aberrant nuclear morphologies, as presented in (a), were scored in individual control, p300-, CBP-, or P/CAF-depleted HeLa cells. (c) Aberrant nuclear morphologies were scored in p300-, CBP-, or P/CAF-depleted HCT116 cells. (d) Control (HeLa Con) and knockdown cells stably expressing p300 (HeLa p300KD) or CBP (HeLa CBPKD) were immunostained with anti-p300 or anti-CBP antibodies and Hoechst dye. p300- and CBP-depleted cells clearly show aberrant nuclear morphologies, multinucleation, and/or aneuploidy
To examine the effect of constitutively inhibiting HAT expression, we transfected HeLa cells with shRNA against p300 or CBP and then selected for stably transformed p300 knockdown (p300KD) and CBP knockdown (CBPKD) cells. The levels of p300 and CBP in the HAT-adapted p300KD and CBPKD clones (i.e., those that escaped cell death and/or mitotic catastrophe) were reduced by approximately 75 and 60%, respectively, relative to their initial levels in control cells (Figure 4d). Chromosome staining clearly showed widespread multinucleation and aneuploidy in p300KD and CBPKD cells, whereas the morphology of chromosomes in control cells was normal (Figure 4d). Consistent with this, flow cytometric analysis using HeLa cells transfected with p300 shRNA, CBP shRNA, or P/CAF shRNA showed a significant increase in the population of aneuploid cells (approximately 6.5% in control cell lines, 14.3% in p300-depleted cell lines, 14.5% in CBP-depleted cell lines, and 17.23% in P/CAF-depleted cell lines) (Supplementary Figure S2). These data suggest that adaptation to HAT depletion induces multinucleation and aneuploidy in cancer cells.
Depletion of HAT in human primary and adult stem cells induced mitotic catastrophe
As cancer cells frequently have defective mitotic cell cycle regulation, they may be particularly prone to HAT depletion and become multinucleated and aneuploid. In addition, the molecular events that lead to a loss of HAT in noncancerous and nonimmortalized cells are not well understood. To address these issues, we expanded our study to include an investigation of the role of HAT in noncancerous human primary cells, including adult stem cells (amniocytes)14 and processed lipoaspirate (PLA) cells.15 We infected freshly isolated amniocytes with a recombinant adenovirus carrying GFP (rAd-GFP), p300 shRNA (rAd-p300 shRNA), CBP shRNA (rAd-CBP shRNA), or P/CAF shRNA (rAd-P/CAF shRNA) and then analyzed the cells by immunoblotting with antibodies against p300, CBP, and P/CAF to determine the knockdown efficiencies (Figure 5a). The recombinant viruses significantly reduced the p300, CBP, or P/CAF levels when compared with cells that were infected with a control rAd-GFP adenoviral construct. Interestingly, live-cell imaging of individual amniocytes showed significant growth retardation and a gradual increase in cell death in HAT shRNA-infected-amniocytes when compared with rAd-GFP-infected control amniocytes. Treatment with the antimicrotubule agent, taxol, produced interesting changes in the morphology of amniocytes, which tended to flatten and shrink (Figure 5b). In addition, treatment with taxol consistently increased the susceptibility of the cells to mitotic catastrophe followed by cell death in HAT-depleted amniocytes, whereas taxol-treated control amniocytes were arrested in mitosis and were resistant to cell death (Figure5b and Supplementary Figure S3).
Depletion of HAT in noncancerous human primary adult stem cells increased their susceptibility to mitotic catastrophe. (a and b) Isolated human primary amniocytes (AF9-1) were infected with a recombinant adenovirus expressing p300 shRNA (rAd-p300 shRNA), CBP shRNA (rAd-CBP shRNA), P/CAF shRNA (rAd-P/CAF shRNA), or GFP (rAd-GFP) as a control. Infected amniocytes were analyzed by immunoblotting with antibodies against p300, CBP, and P/CAF (a). Infected amniocytes were further cultured in the presence or absence of taxol for 24 or 48 h, after which they were digitally imaged by photomicroscopy (b). (c) Infected amniocytes were immunostained with anti-MPM2 and anti-active caspase 3 (caspase 3a) antibodies. Data represent the average number of cells that were positive for both phospho-H3 and caspase 3a on the basis of three independent experiments; ⩾200 cells/sample were counted in each experiment. (d) Processed lipoaspirate (PLA) cells, adipose-derived stem cells, were infected with rAd-GFP or rAd-p300 shRNA. Infected cells were further cultured in the presence or absence of taxol for 24 h and then digitally imaged
To examine the frequency of mitotic catastrophe following HAT depletion, we infected amniocytes with the indicated HAT shRNA adenoviral constructs and then immunostained them for caspase 3a and the mitotic marker, MPM2 (Figure 5c). Only approximately 3.2% of the control rAd-GFP-infected amniocytes were positive for both MPM2 and caspase 3a. However, depletion of HAT significantly increased the frequency of MPM2/caspase 3a double-positive cells. Indeed, following knockdown of p300, CBP, or P/CAF, the fraction of cells positive for both markers was approximately 12.4, 15.4, and 15.9%, respectively.
We also infected PLA cells with rAd-GFP or rAd-p300 shRNA and then cultured them in the presence of taxol. Similar to the amniocytes, taxol treatment resulted in increased susceptibility to mitotic catastrophe and subsequent cell death in p300-depleted PLA cells, whereas taxol-treated control PLA cells were apparently resistant to cell death (Figure 5d). Together, these data suggest that depleting HAT expression in human primary and adult stem cells induced mitotic catastrophe.
Depletion of HAT augmented H2AX phosphorylation in mitotic cells
Histone H2AX is rapidly phosphorylated at the Ser139 residue in response to ionizing radiation or other agents that introduce DNA double-strand breaks (DSBs; see review).16 This phosphorylated H2AX, called γH2AX, forms clusters at each DSB site, which enables visualization of the nuclear foci using γH2AX antibody.17 Recent studies have provided important new evidence that H2AX is phosphorylated independently of DSB and DNA damage signaling, and that this phosphorylation is required for proper mitotic chromatin condensation to occur.18, 19 Ataxia telangiectasia-mutated (ATM)-mediated H2AX phosphorylation may also be coupled to mitotic catastrophe.20, 21 Moreover, it has been reported that H2AX phosphorylation induces DNA fragmentation and degradation through activated DNase, which indicates that H2AX phosphorylation is a prerequisite for the cell death process.22, 23 However, the biological function of mitotic H2AX phosphorylation is still not known.
We evaluated the effect of HAT depletion on the levels of phosphorylated H2AX and H2AX foci formation in HeLa cells that had been treated with taxol (Figure 6). HeLa cells tansfected with control shRNA showed a slight increase in H2AX phosphorylation in condensed (pro)metaphase chromosomes following treatment with taxol, which is consistent with the results of previous studies.18 Conversely, treatment with taxol led to markedly increased levels of H2AX phosphorylation in p300-, CBP-, or P/CAF-depleted cells after 24 h (Figure 6a). We confirmed these results by evaluating the formation of γH2AX. We could readily detect γH2AX foci in control shRNA-transfected HeLa cells following treatment with taxol, but the intensity and number of foci were greater in p300-, CBP-, or P/CAF-depleted HeLa cells (Figure 6b and c). It is most likely that the disorganized and abnormal mitotic chromosome morphology generated by HAT depletion (Figure 4a) is more susceptible to H2AX phosphorylation. Notably, we observed some γH2AX foci in HAT-depleted mitotic phase cells, even in the absence of the microtubule inhibitor (Figure 6c). When combined with our earlier data shown in Figures 3 and 5, these results show that depletion of HAT induced mitotic catastrophe and increased cellular susceptibility to mitotic catastrophe in response to a microtubule inhibitor. Overall, it is worthwhile to investigate whether the phosphorylation of H2AX could be one of the signaling processes of mitotic catastrophe.
H2AX phosphorylation and foci formation induced by HAT depletion. (a) HeLa cells were transfected with pS, pS-p300 shRNA, -CBP shRNA, or -P/CAF shRNA, and then treated with taxol. Cells were harvested at the indicated times and immunoblotted with antibodies specific for H2AX phosphorylated at Ser 139 (γH2AX) and actin. (b) HeLa cells were transfected as described in (a) and then immunoblotted with anti-phospho-H2AX (γH2AX) and anti-MPM2 antibodies and CREST serum. DNA was visualized using Hoechst dye. γH2AX foci are indicated by arrowheads. (c) HeLa cells were transfected with control shRNA (pS), p300 shRNA, CBP shRNA, or P/CAF shRNA, after which the cells were treated with taxol. Cells were immunostained as described in (b). Data represent the frequency of γH2AX foci formation in MPM2-positive mitotic cells. At least 1200 cells/sample were counted per experiment
Depletion of HATs resulted in the inhibition of condensin subunit expression
In our initial attempt to understand the role that HAT proteins play in mitotic chromatin condensation and decondensation, we showed that the levels of histone acetylation were maximal during the prophase-to-metaphase period of mitosis, during which time DNA staining showed that cells exhibited normal condensed mitotic chromosomes (Supplementary Figure S4). These results indicated an unexpected correlation between mitotic chromosome condensation and histone acetylation.
At present, the method by which cells govern the processes of chromosome condensation or decondensation during the mitotic phase of the cell cycle is not well understood. Recent studies have suggested that the condensin complex contributes directly to mitotic chromosome assembly and condensation.23 During prophase, chromosome condensation is associated with the progressive folding of large-scale chromatin fibers, a process that is accompanied by the redistribution of topoisomerase II and condensin subunits (structural maintenance of chromosome 2 (SMC2) and CAP-E) within the chromosome.24, 25 These observations strongly suggest that recruiting condensin to chromosomes, where it most likely acts as an intramolecular DNA crosslinker, plays a critical role in mitotic chromosome condensation. Condensin subunits consistently associate with a peripheral region of prophase chromatin, after which they become progressively enriched at the central axis of metaphase chromatids.26, 27 In addition, the condensin complex may convert relaxed circular DNA into positively supercoiled DNA.28, 29 The emerging view is that chromosome condensation and compaction during mitosis are multifunctional processes that include the dynamic distribution and localization of condensin.
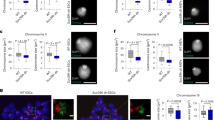
To examine the effect of HAT protein depletion on the expression of condensin subunits, we transfected HeLa cells with control pSuper or the pSuper-p300 shRNA, -CBP shRNA, or -P/CAF shRNA and then treated the transfected cells with taxol (Figure 7a). In the shRNA-transfected cells, the p300, CBP, or P/CAF protein levels were substantially depleted and the hCAP-D3 levels were markedly reduced, even in the absence of taxol treatment. Furthermore, the hCAP-D3 levels in the shRNA-transfected cells were almost undetectable 36 h post-treatment. However, the levels of hCAP-D3 in the control shRNA-transfected cells showed only a slight reduction at 24 h after taxol treatment. These results show that HAT activity affects the levels of condensin subunit D3. Conversely, the abundance of another condensin subunit, hCAP-G, was not affected by HAT depletion, although it was rapidly attenuated 36 h after treatment with taxol, regardless of the HAT protein levels. To compare the levels of hCAP-D3 mRNA between the control and HAT-depleted cells, we extracted the total RNA and then amplified the mRNA by quantitative real-time PCR (qRT-PCR) (Figure 7b). However, we did not observe any significant changes in the hCAP-D3 mRNA levels in response to HAT depletion.
Reduction of condensin subunit expression in HAT-depleted cells. (a) HeLa cells were transfected with control shRNA (pS), pS-p300 shRNA, -CBP shRNA, or -P/CAF shRNA, and then treated with taxol. At the indicated times, the cells were harvested and immunoblotted with anti-p300, anti-CBP, anti-P/CAF, anti-hCAP-D3, anti-hCAP-G, and antiactin antibodies. Arrows indicate the markedly reduced levels of hCAP-D3 in HAT-depleted cells. (b) Quantitative real-time PCR (qRT-PCR) conducted to measure the levels of hCAP-D3 mRNA. Total RNA was extracted from the transfected cells as described in (a), and mRNA was amplified by qRT-PCR as described in the Materials and Methods
Depletion of hCAP-D3 led to mitotic delay and mitotic catastrophe
To determine if depletion of hCAP-D3 led to an aberrant mitotic duration and/or mitotic catastrophe as seen in the HAT-depleted cells, we transfected HeLa cells with control luciferase shRNA or hCAP-D3 shRNA and then applied nocodazole treatment to synchronize the transfected cells (Figure 8a and b). Following release from the nocodazole treatment (NR), the cells were harvested at various time points and then analyzed by immunoblotting (Figure 8a) and flow cytometry (Figure 8b). Interestingly, hCAP-D3-depleted cells exhibited a significant increase in the levels of the mitotic marker proteins, cyclin B1 and phospho-H3, at 1–2 h NR. In addition, the levels of Mad2 were much more stable in hCAP-D3-depleted cells over the entire course of treatment. Similarly, flow cytometric analysis using these cells revealed that hCAP-D3-depleted cells displayed a significant delay in the exit from mitosis when compared with the control cells. To further examine the morphological phenotype of cells lacking hCAP-D3, we cotransfected HeLa cells with an expression plasmid encoding H2B-GFP and hCAP-D3 shRNA. Time-lapse microscopic analysis revealed that mitosis was prolonged in the hCAP-D3-depleted cells (Figure 8c). Indeed, the average mitotic duration of hCAP-D3-depleted cells was about 99 min, whereas that of the control cells was 63 min (Figure 8d). Moreover, a high percentage of cells exhibited features of mitotic catastrophe following transient depletion of hCAP-D3 (from 8.8% in the control shRNA-transfected cells to 23.2% in the treated cells) (Figure 8e). Taken together, these data clearly show that a loss of hCAP-D3 resulted in a severe delay in the exit from mitosis and mitotic catastrophe, as shown in the HAT-depleted cells.
Depletion of hCAP-D3 leads to an aberrant delay in mitotic exit and mitotic catastrophe. (a and b) HeLa cells were transfected with control luciferase (Luc) shRNA or hCAP-D3 shRNA, after which the cells were synchronized by nocodazole treatment (N; 100 ng/ml) or nocodazole treatment and then released into the cell cycle (NR). Cells were harvested at the indicated times for immunoblotting with anti-hCAP-D3, anti-BubR1, anti-cyclin B1, anti-Bub3, anti-Mad2, anti-securin, anti-Phospho-H3, and antiactin antibodies (a). At the indicated times, cells were stained with propidium iodide and then analyzed by flow cytometry (b). (c) HeLa cells were transfected with a plasmid encoding H2B-GFP and control luciferase shRNA or hCAP-D3 shRNA. Cells were then cultured, after which mitotic progression was imaged by time-lapse microscopy. Times from the breakdown of the nuclear envelope to complete chromosome separation are indicated. Representative time-lapse images of cells transfected with each shRNA are shown. (d and e) At least 40 cells/sample were counted per experiment; data show the mitotic time duration (d) and mitotic catastrophe population (e)
In summary, our results showed that depletion of HAT in cancer cells resulted in severe mitotic defects that led to mitotic catastrophe or, alternatively, to multinucleation and aneuploidy in a subpopulation of cells that adapted to HAT depletion. However, the predominant response to HAT depletion in noncancerous cells and adult stem cells was mitotic catastrophe, not adaptation to mitotic defects. It is important to note that mitotic catastrophe in HAT-depleted cells appeared to be mediated by a signaling process involving H2AX phosphorylation. Therefore, these results provide novel molecular evidence that HAT proteins monitor mitotic chromatin integrity.
Discussion
Histone acetylation destabilizes chromatin fibers and is sufficient to promote decondensation of chromatin in interphase cells.1, 30 During mitosis, chromosome condensation is crucial to ensure accurate segregation of chromosomes. On the basis of evidence at present, however, it is reasonable to infer that histone acetylation and deacetylation may have only a limited impact on decondensation and condensation of mitotic chromosomes, respectively.27, 31 Nevertheless, it is clear that histone hyperacetylation induced by pharmacological HDAC inhibitor treatment leads to severe defects in mitotic progression, although the precise underlying mechanism remains unknown.3 In this study, we examined the role of HAT proteins in mitotic chromatin remodeling and mitotic progression using shRNA-mediated depletion of three representative HATs. We found that the loss of HAT induced mitotic catastrophe and activated the process of mitotic cell death, whereas adaptation to HAT depletion, which occurred in a subpopulation of the cancer cells, led to multinucleation and aneuploidy. These findings support the concept that HATs act as ‘gatekeepers’ in regulating the integrity of mitotic chromosomes, explaining why the predominant response to HAT depletion is mitotic catastrophe.
Although the molecular mechanisms that regulate mitotic catastrophe by HAT proteins require further clarification, mitotic catastrophe induced by HAT depletion appears to stem from the activation of H2AX phosphorylation. Recent studies have revealed that phosphorylation of H2AX increases DNA cleavage by caspase-activated DNase, which induces DNA fragmentation,22 indicating that H2AX phosphorylation is required for cells to undergo apoptotic cell death. Importantly, additional studies have found that H2AX phosphorylation in mitotic chromosomes occurs independently of DSB and DNA damage signaling, and this phosphorylation appears to be required for proper mitotic chromatin condensation.18, 19 Therefore, depletion of HAT proteins during mitosis leads to highly aberrant condensation of mitotic chromosomes, which may yield structures that are more prone to inducing H2AX phosphorylation signaling.
The present model behind how depletion of HAT proteins activates phosphorylation of H2AX at the mitotic phase, even in the absence of ionizing radiation or DNA damage, is as follows. At the beginning of mitosis, the dynamic changes in chromatin structure associated with the formation of condensed chromosomes promote ATM kinase activity, resulting in phosphorylation of H2AX in the mitotic phase. It is noteworthy that cells exposed to mildly hypotonic buffers or chromatin-modifying drugs that do not induce DSBs show rapid autophosphorylation of ATM,32, 33 indicating that changes in chromatin structure, independently of DSBs or DNA damage, could stimulate ATM autophosphorylation.18 Therefore, it seems that H2AX phosphorylation induced by aberrant chromosome condensation in HAT-depleted mitotic cells could be a key in the signaling pathway that leads to mitotic catastrophe. Such a mechanism prevents cells from adapting to aberrant chromosome architecture and/or chromosome abnormalities.
The condensation of chromosomes during mitotic entry is an essential prerequisite for successful chromosome segregation at cell division. An intensive examination of these events revealed that mitotic chromosome condensation is a complicated, multistep process: primary condensation converts diffuse chromatin into discrete linear chromosomes. After a plateau phase, secondary condensation compacts the curvilinear chromosomes to form shorter, bar-shaped structures.34 How cells govern the mitotic chromosome condensation and decondensation processes is still a challenging question, but strong evidence suggests that condensin subunits and chromosome arm-binding kinesins are essential components of mitotic chromosome condensation and compaction.34 For example, heterodimeric SMC2–SMC4 condensin subunits bind to dsDNA and are important in chromosome assembly during mitosis.35 Other recent studies have shown that non-SMC condensin subunits can physically compact DNA in an ATP-hydrolysis-dependent manner, and that they also have the ability to organize DNA supercoils into an ordered assembly through a wrapping mechanism.28, 29 These models are consistent with recent reports that condensin subunits first associate with peripheral regions of prophase chromatin and then become progressively enriched in the central axis of metaphase chromatids.26, 27 Taken together, these observations strongly support the idea that condensin directly contributes to mitotic chromosome assembly and condensation.
In this study, we examined the effect of HATs on the expression of non-SMC condensin subunits, such as CAP-D3 and CAP-G, which share a highly degenerate repeating motif known as HEAT repeat. This motif is required for establishing and maintaining mitotic chromosome structure.36 Interestingly, the shRNA-mediated HAT depletion marked reduced hCAP-D3 expression. This result yields important new evidence directly implicating HAT proteins in the regulation of condensin subunit expression. However, we do not exclude the possibility that CAP-D3 protein might be degraded during the process of cell death. In addition, the HAT transcription cofactor, transformation/transcription domain-associated protein (Trrap), reportedly regulates the expression of the spindle checkpoint proteins, Mad1 and Mad2; furthermore, loss of Trrap leads to mitotic checkpoint defects.37 Similarly, it is also possible that HAT proteins regulate the expression of genes that are required for mitotic cell cycle and checkpoint controls, at least in part (Figure 1d).
Some overlap exists between HDAC inhibitor-induced histone hyperacetylation and mitotic cell cycle control, but regulating mitotic chromatin integrity by HDAC inhibition is apparently not a direct consequence of changes in molecular signaling that result from histone hyperacetylation. Previous work has revealed clues as to the molecular actions underlying HDAC inhibition-induced histone hyperacetylation that control mitotic cell cycle and/or mitotic chromatin remodeling. Initially, HDAC inhibitors were designed to block cell proliferation by inhibiting signaling pathways and altering gene expression; they also induced and/or sensitized cells to apoptotic cell death.38 Others have proposed that HDAC inhibitors may cause improper localization of mitotic checkpoint proteins and induce aberrant mitotic arrest and/or cytokinesis failure.4, 5 HDAC inhibitors, depending on their concentration, time of administration, duration of treatment, and/or specific genetic or epigenetic cellular context, may increase the susceptibility of cancer cells to aneuploidy.38, 39 These distinctions can be important in identifying cellular determinants of sensitivity to HDAC inhibitors. For example, through HAT depletion or inactivation, HDAC may selectively sensitize cancer cells to the proapoptotic effects of antimitotic cancer drugs, such as paclitaxel or vinca alkaloids.
In summary, depletion of HAT in cancer cells resulted in severe mitotic defects that led to mitotic catastrophe or, alternatively, to multinucleation and aneuploidy in cells that adapted to HAT. In contrast, human primary adult stem cells responded to HAT depletion by undergoing mitotic catastrophe rather than by adapting to the mitotic defect. Moreover, mitotic catastrophe induced by HAT depletion appears to be mediated by H2AX phosphorylation, which ultimately leads to DNA fragmentation. These data provide new evidence that HATs function as ‘gatekeepers’ in maintaining mitotic chromosome stability and integrity, rather than serving to regulate mitotic chromosome condensation.
Materials and Methods
Cell culture and generation of stable knockdown cells
HeLa and HCT116 cells were cultured at 37 °C in a humidified 5% CO2 incubator. HeLa cells were purchased from the American Type Culture Collection and grown in Dulbecco's modified Eagle's medium (DMEM; Hyclone) containing 10% fetal bovine serum (FBS; Hyclone). Amniocytes (AF9-1) were obtained from human amniotic fluid supernatants. Informed consent was obtained, and the cells were used in accordance with the IRB guidelines of Kangbuk Samsung Hospital, Sungkyunkwan University School of Medicine. AF9-1 cells were expanded by adherent culture in minimal essential medium (GIBCO BRL) containing 20% FBS and 4 ng/ml basic fibroblast growth factor (bFGF, GIBCO-BRL). PLA cells were isolated from the fatty portion of liposuction aspirates, plated in gelatin-coated culture dishes, and cultured as described previously.15 Stable knockdown cell lines of p300 (p300KD) and CBP (CBPKD) were generated by transfecting HeLa cells with pSuper.puro-p300 and pSuper.puro-CBP shRNAs, respectively (see plasmid descriptions below), and culturing them in the presence of puromycin (10 μg/ml). Puromycin-resistant colonies were clonally selected and screened by immunoblotting and immunofluorescence microscopy, using anti-p300 (Santa Cruz), anti-CBP (Abcam), and P/CAF (Santa Cruz) antibodies to confirm suppression of the targeted HATs.
Plasmids, adenoviral vectors, and transfection
Oigonucleotides encoding shRNAs against p300, CBP, P/CAF, and hCAP-D3 were synthesized and inserted into the pSuper vector or pSuper.puro vectors, which contain an H1 promoter and T5 terminal sequences (Oligoengine). Specific shRNA sequences used: p300 (p300 shRNA no. 1 5′-gatccccgcacaaatgtctagttcttttcaagagaaagaactagacatttgtgctttttggaaa-3′/5′- agcttttccaaaaagcacaaatgtctagttctttctcttgaaaagaactagacatttgtgcggg-3′ and p300 shRNA no. 2 5′-gctccccgcagctcaaccatccactattcaagagatagtggatggttgagctgctttttggaaa-3′/5′-agcttttccaaaaagcagctcaaccatccactatctcttgaatagtggatggttgagctgcggg-3′), CBP (CBP shRNA no. 1 5′-gatccccgcagcagccagcattgatattcaagagatatcaatgctggctgctgctttttggaaa-3′/5′-agcttttccaaaaagcagcagccagcattgatatctcttgaatatcaatgctggctgctgcggg-3′ and CBP shRNA no. 2 5′-gatccccctgatgagctgatacccaatgttcaagagacattgggtatcagctcatcagtttttggaaa-3′/5′ agcttttccaaaaactgatgagctgatacccaatgtctcttgaacattgggtatcagctcatcagggg-3′) and P/CAF (P/CAF shRNA no. 1 5′-gatccccgctcgtctttgacccgaaattcaagagatttcgggtcaaagacgagctttttggaaa-3′/5′-agcttttccaaaaagctcgtctttgacccgaaatctcttgaatttcgggtcaaagacgagcggg-3′ and P/CAF-shRNA no. 2 5′-gatccccggagccactttaatgggatttcaagagaatcccattaaagtggctcctttttggaaa-3′/5′-agcttttccaaaaaggagccactttaatgggattctcttgaaatcccattaaagtggctccggg-3′), hCAP-D3 (hCAP-D3 shRNA no. 1 5′-gatccccctggatttcacagagactgttttcaagagaaaccgtctctgtgaaatccagtttttggaaa-3′/5′-agcttttccaaaaactggatttcacagagactgtttctcttgaaaaccgtctctgtgaaatccagggg-3′ and hCAP-D3 shRNA no. 2 5′-gatccccgcagagatcatagagactgttttcaagagaaacagtgtctatgatctctgctttttggaaa-3′/5′-agcttttccaaaaagcagagatcatagagactgtttctcttgaaaacagtgtctatgatctctgcggg-3′). For adenoviral vector preparation, DNA fragments containing the H1 promoter, target-specific shRNA, and a T5 sequence were excised from the pSuper-based plasmids and inserted into pShuttle vectors (Quantum). Adenoviral vectors were subsequently generated by recombination of the pShuttle plasmids encoding shRNAs with pAdeasy 1 in Escherichia coli BJ5183. The resulting recombinant DNAs were digested with PacI and transfected into 293A cells. Recombinant adenoviruses were purified and titered using a 293A cell plaque-forming assay. For transient transfections, cells were electroporated using a microporator (Digital Biotechnology).
Synchronization and cell cycle analysis
HeLa cells were synchronized by treatment with nocodazole (100 ng/ml) for 16 h followed by release, to generate cells enriched in the post-mitotic and G1 phases of the cell cycle. Cells were harvested at various times after synchronization and analyzed for DNA content by flow cytometry as follows: Synchronized cells were harvested, fixed in 70% (vol/vol) ethanol, washed with cold-phosphate-buffered saline (PBS), and stained with 40 μg/ml propidium iodide (PI) in the presence of 50 μg/ml RNase A for 30 min at room temperature. Cellular DNA (10 000 cells/sample) was analyzed with a Becton Dickinson FACScan cytometer using the CellQuest (Clontech) and WinMDI 2.8 (http://facs.scripps.edu/software.html) software packages.
Immunoblotting analysis and immunofluorescence staining
For immunoblot analysis, synchronized cells prepared as described above were harvested by scraping, washed twice in cold PBS, and lysed in nuclear extraction buffer (20 mM HEPES (pH 7.6), 20% glycerol, 250 mM NaCl, 1.5 mM MgCl2, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and protease inhibitor cocktail (Roche)). Protein concentrations in the nuclear extracts were determined using the Bio-Rad Protein Assay with bovine serum albumin as a standard. Samples containing equal amounts of protein were separated by SDS-PAGE, transferred to nitrocellulose membranes, blocked, and probed with antibodies specific for p300 (Santa Cruz), CBP (Abcam), P/CAF (Santa Cruz), H2AX phosphorylated at Ser139, hCAP-D3, hCAP-G (Bethy), Mad2, BubR1 (BD Biosciences, Pharmingen), Securin (Zymed), phospho-histone H3 at ser10 (P-H3; Upstate Biotechnology), cyclin B1 (Santa Cruz) and actin (Sigma) antibodies. For immunofluorescence assays, cells were cultured on 18-mm coverslips and fixed in 5% formaldehyde for 10 min. Fixed cells were washed, permeabilized with 0.1% Triton X-100 in PBS, and incubated at room temperature for 2 h with anti-active caspase 3 (Caspase 3a; Cell Signaling Technology), anti-MPM2 (Abcam), anti-phospho-histone H3 at Ser10 (P-H3; Upstate Biotechnology) antibodies, where indicated. The cells were then washed and incubated for 1 h with goat anti-mouse IgG conjugated to Alexa Fluor 488 or goat anti-Rabbit IgG conjugated to Alexa Fluor 568 (Molecular Probes). After incubation with secondary antibodies, the cells were washed, stained with Hoechst dye to visualize DNA, and viewed under a confocal microscope (Zeiss 510 Meta) or fluorescence microscope (Carl Zeiss Axiophoto II microscope).
Live cell imaging
Chromosomes were visualized in HeLa cells or amniocytes cotransfected with an expression plasmid encoding green GFP-H2B or red fluorescent protein RFP-H2B and pSuper-p300 shRNA no. 2 (or pSuper-CBP shRNA no. 1 or P/CAF shRNA no. 1 or hCAP-D3 shRNA no. 1). Transfected cells, maintained in DMEM containing 10 or 20% FBS and 4 ng/ml bFGF (Invitrogen), were imaged in ⩾T 0.15 mm dishes. Estimates of mitosis duration and analysis of mitotic catastrophe were based on 30-ms exposures taken every 3 min for 18 h using an LSM500 META confocal microscope (Carl Zeiss) with a × 40 NA0.75 objective, or using an Axiovert 200 M microscope containing a Zeiss AxioCam HRm (Carl Zeiss).
qRT-PCR of hCAP-D3 mRNA
The RNeasy RNA extraction kit (Qiagen) was used to extract total RNA. A total of 1 μg of RNA was reverse-transcribed and used to generate first-strand cDNA (First Strand cDNA kit, Roche). After quantification by fluorometry, 5 ng of cDNA was subjected to 40 cycles of real-time PCR in a LightCycler (Roche), according to the manufacturer's instructions. Intron-spanning cDNA primers for the reference gene, GAPDH, were used to confirm adequate cDNA normalization. The primers used for hCAP-D3 mRNA detection were residues 714–832 (5′-TTTGGAGCAAGAGTCGAATG-3′ and 5′-GTGGTGTCCCGATGTAACTG-3′) and residues 147–279 (5′-CCTGAAGACTGTGCTGGAGA-3′ and 5′- CTCTGATGCCAGCTGTTTGT-3′).
Abbreviations
- HDAC:
-
histone deacetylases
- HAT:
-
histone acetyltransferase
- phospho-H3:
-
phosphorylated histone H3
- KD:
-
knockdown
- GFP:
-
green fluorescence protein
- rAd:
-
recombinant adenovirus
- PLA:
-
processed lipoaspirate
- SMC:
-
structural maintenance of chromosomes
- NR:
-
nocodazole treatment
- ATM:
-
ataxia telangiectasia-mutated
- Trrap:
-
transformation/transcription domain-associated protein
- Luc:
-
luciferase
- qRT-PCR:
-
quantitative real-time PCR
- DMEM:
-
Dulbecco's modified Eagle's medium
- FBS:
-
fetal bovine serum
- bFGF:
-
basic fibroblast growth factor
- PBS:
-
phosphate-buffered saline
References
Ura K, Kurumizaka H, Dimitrov S, Almouzni G, Wolffe AP . Histone acetylation: influence on transcription, nucleosome mobility and positioning, and linker histone-dependent transcriptional repression. EMBO J 1997; 16: 2096–2107.
Lee KK, Workman JL . Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol 2007; 8: 284–295.
Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F . Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell 2003; 14: 3821–3833.
Shin HJ, Baek KH, Jeon AH, Kim SJ, Jang KL, Sung YC et al. Inhibition of histone deacetylase activity increases chromosomal instability by the aberrant regulation of mitotic checkpoint activation. Oncogene 2003; 22: 3853–3858.
Stevens FE, Beamish H, Warrener R, Gabrielli B . Histone deacetylase inhibitors induce mitotic slippage. Oncogene 2008; 27: 1345–1354.
Taddei A, Maison C, Roche D, Almouzni G . Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol 2001; 3: 114–120.
Shin HJ, Baek KH, Jeon AH, Park MT, Lee SJ, Kang CM et al. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 2003; 4: 483–497.
Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM . Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 2004; 6: 232–237.
Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 2001; 409: 355–359.
Kops GJ, Weaver BA, Cleveland DW . On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 2005; 5: 773–785.
Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ 2008; Oct: 10.
Ricci MS, Zong WX . Chemotherapeutic approaches for targeting cell death pathways. Oncologist 2006; 11: 342–357.
Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G . Cell death by mitotic catastrophe: a molecular definition. Oncogene 2004; 23: 2825–2837.
Tsai MS, Lee JL, Chang YJ, Hwang SM . Isolation of human multipotent mesenchymal stem cells from second-trimester amniotic fluid using a novel two-stage culture protocol. Hum Reprod 2004; 19: 1450–1456.
Yoshimura K, Shigeura T, Matsumoto D, Sato T, Takaki Y, Aiba-Kojima E et al. Characterization of freshly isolated and cultured cells derived from the fatty and fluid portions of liposuction aspirates. J Cell Physiol 2006; 208: 64–76.
Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W . Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev 2002; 12: 162–169.
Rogakou EP, Boon C, Redon C, Bonner WM . Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol 1999; 146: 905–916.
Ichijima Y, Sakasai R, Okita N, Asahina K, Mizutani S, Teraoka H . Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem Biophys Res Commun 2005; 336: 807–812.
Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 2003; 4: 497–508.
Rainey MD, Black EJ, Zachos G, Gillespie DA . Chk2 is required for optimal mitotic delay in response to irradiation-induced DNA damage incurred in G2 phase. Oncogene 2008; 27: 896–906.
Niida H, Tsuge S, Katsuno Y, Konishi A, Takeda N, Nakanishi M . Depletion of Chk1 leads to premature activation of Cdc2-cyclin B and mitotic catastrophe. J Biol Chem 2005; 280: 39246–39252.
Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell 2006; 23: 121–132.
Hirota T, Lipp JJ, Toh BH, Peters JM . Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 2005; 438: 1176–1180.
Kireeva N, Lakonishok M, Kireev I, Hirano T, Belmont AS . Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J Cell Biol 2004; 166: 775–785.
Belmont AS . Mitotic chromosome structure and condensation. Curr Opin Cell Biol 2006; 18: 632–638.
Maeshima K, Laemmli UK . A two-step scaffolding model for mitotic chromosome assembly. Dev Cell 2003; 4: 467–480.
Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC . Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 2003; 5: 323–336.
Bazett-Jones DP, Kimura K, Hirano T . Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol Cell 2002; 9: 1183–1190.
Strick TR, Kawaguchi T, Hirano T . Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 2004; 14: 874–880.
Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A . Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol 2005; 25: 7216–7225.
McManus KJ, Hendzel MJ . The relationship between histone H3 phosphorylation and acetylation throughout the mammalian cell cycle. Biochem Cell Biol 2006; 84: 640–657.
McGowan CH, Russell P . The DNA damage response: sensing and signaling. Curr Opin Cell Biol 2004; 16: 629–633.
Bakkenist CJ, Kastan MB . DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003; 421: 499–506.
Hirano T . At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 2006; 7: 311–322.
Yoshimura SH, Hizume K, Murakami A, Sutani T, Takeyasu K, Yanagida M . Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr Biol 2002; 12: 508–513.
Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 1999; 274: 25543–25549.
Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z . HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J 2004; 23: 4824–4834.
Burgess A, Ruefli A, Beamish H, Warrener R, Saunders N, Johnstone R et al. Histone deacetylase inhibitors specifically kill nonproliferating tumour cells. Oncogene 2004; 23: 6693–6701.
Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW . Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 2007; 11: 25–36.
Acknowledgements
We thank Dr. Seong Tae Kim for comments on the manuscript. This study was supported by research grants from the Korea Research Foundation (KRF-2006-C00324), the 21C Frontier Functional Human Genome Project from the Ministry of Science and Technology in Korea (FG07-21-01), and the Korea Science and Engineering Foundation through the Rheumatism Research Center (RII-2002-098-05003, 2007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Blagosklonny
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Rights and permissions
About this article
Cite this article
Ha, GH., Kim, HS., Lee, C. et al. Mitotic catastrophe is the predominant response to histone acetyltransferase depletion. Cell Death Differ 16, 483–497 (2009). https://doi.org/10.1038/cdd.2008.182
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.182
Keywords
This article is cited by
-
SIRT1 regulates mitotic catastrophe via autophagy and BubR1 signaling
Molecular and Cellular Biochemistry (2022)
-
Thymine DNA glycosylase as a novel target for melanoma
Oncogene (2019)
-
Cyclizing-berberine A35 induces G2/M arrest and apoptosis by activating YAP phosphorylation (Ser127)
Journal of Experimental & Clinical Cancer Research (2018)
-
CBP and P300 regulate distinct gene networks required for human primary myoblast differentiation and muscle integrity
Scientific Reports (2018)
-
Linking site-specific loss of histone acetylation to repression of gene expression by the mycotoxin ochratoxin A
Archives of Toxicology (2018)