Abstract
Stimulation of death receptors activates the extrinsic apoptotic signaling pathway that leads to cell death. Although many steps of this apoptotic signaling cascade are known, few mechanisms that counterbalance the death signal have been described. We identified an antiapoptotic protein complex associated with death receptors that contains glycogen synthase kinase-3 (GSK3), DDX3 and cellular inhibitor of apoptosis protein-1 (cIAP-1). GSK3, DDX3 and cIAP-1 are associated in cells with each other and with death receptors. Blocking the actions of GSK3 or DDX3 potentiated caspase-3 activation induced by stimulation of four different death receptors in several types of cells. GSK3 restrained apoptotic signaling by inhibiting formation of the death-inducing signaling complex and caspase-8 activation. Stimulated death receptors surmount the antiapoptotic complex by causing GSK3 inactivation and cleavage of DDX3 and cIAP-1 to enable progression of the apoptotic signaling cascade, but the antiapoptotic complex remains functional in cancer cells resistant to death receptor stimulation, a resistance that is overcome by GSK3 inhibitors. Thus, an antiapoptotic complex of GSK3, DDX3 and cIAP-1 caps death receptors, providing a checkpoint to counterbalance apoptotic signaling.
Similar content being viewed by others
Main
The extrinsic apoptotic signaling cascade is a vital process initiated by activation of death receptors that causes cell death. These death receptors are members of the tumor necrosis factor (TNF) receptor family that contain conserved intracellular death domains that are critical for the initiation of apoptotic signaling.1 The most common death receptors capable of inducing apoptosis are Fas (CD95/Apo1), TNF-R1 (p55/CD120a), TNF-related apoptosis-inducing ligand receptor-1 (TRAIL-R1/DR4) and TRAIL-R2 (DR5/Apo2/TRICK2/KILLER). Stimulation of these death receptors causes receptor trimerization, followed by recruitment of FADD (Fas associated with death domain protein) and caspase-8 (or caspase-10) to form the death-inducing signaling complex (DISC).2 DISC formation promotes autoactivation of caspase-8/10 and the subsequent activation of effector caspases, primarily caspase-3, -6 and -7, which implement the cell death program.3
Although the basic extrinsic apoptosis signaling pathway has been described in detail, less is known about counteractive mechanisms that abrogate apoptotic signaling by death receptors. Several proteins are known to provide protection from many forms of apoptosis, such as bcl-2, but only few proteins are known that act specifically at death receptors to inhibit apoptosis, such as c-FLIP4 and the cellular inhibitor of apoptosis protein-1 (cIAP-1) at the TNF receptor.5, 6, 7 Another antiapoptotic protein is DDX3, which was recently indicated to impede TRAIL-R2-induced apoptosis in resistant cancer cells.8 Glycogen synthase kinase-3 (GSK3), comprised of GSK3α and GSK3β isoforms, also is antiapoptotic toward death receptor-induced apoptotic signaling.9 Evidence implicating GSK3 in extrinsic apoptotic signaling derives from the early finding that lithium promotes TNF-mediated cytotoxicity10, 11 and the subsequent discovery that lithium directly and selectively inhibits GSK3.12 Tying these two findings together was the discovery that knocking out GSK3β caused mouse embryonic lethality because of TNF hypersensitivity in the liver,13 which provided the key insight that GSK3β inhibits TNF-induced apoptosis. This conclusion was supported by studies showing that lithium potentiated TNF-induced cytotoxicity in mouse embryonic fibroblasts from wild-type mice13 and in hepatocytes.14 Thus, knocking out GSK3β or inhibiting GSK3 with lithium potentiated TNF-induced apoptosis, indicating an antiapoptotic role for GSK3β. Subsequently, GSK3 has been found to be dysregulated in many types of cancers, although how this contributes to cancerous growth remains unclear.15
The limited knowledge about mechanisms that counteract death receptor-induced apoptotic signaling prompted us to examine further the possibilities that GSK3 and DDX3 may antagonize signaling by death receptors to inhibit apoptotic signaling, with a focus on TRAIL-R2-mediated signaling. The results show that the antiapoptotic actions of GSK3 and DDX3 extend to each of the four major death-inducing receptors, that GSK3 inhibits DISC formation and that GSK3 associates with TRAIL-R2, DDX3 and cIAP-1 at the receptor. These findings indicate that GSK3, DDX3 and cIAP-1 form a death-antagonizing signaling complex at death receptors to inhibit apoptotic signaling. Such a complex could ensure maintenance of unstimulated death receptors in a quiescent state and play a role in setting the threshold for death signaling after stimulation, as well as potentially contribute to the development of resistance to death receptor-induced signaling in cancer cells.
Results
GSK3 impedes death receptor-induced caspase-3 activation
Inhibition of GSK3 promoted apoptotic signaling induced by each of the four major death receptors in several cell types. Using the agonistic antibody TRA-8 to specifically stimulate TRAIL-R216 in MDA-MB-231 cells, the selective GSK3 inhibitor lithium12 increased caspase-3 activity by two- to fourfold, with greatest potentiation achieved with low TRA-8 concentrations (Figure 1a). This indicates that GSK3 provides an antiapoptotic check on signaling induced by TRAIL-R2. This apoptosis-potentiating effect of lithium was also evident in cells derived from MDA-MB-231 cells with high resistance to TRAIL-R2-induced apoptosis.8 In resistant MDA-MB-231 cells, caspase-3 activity induced by TRA-8 stimulation of TRAIL-R2 was only ∼20% of parental MDA-MB-231 cells (Figure 1a). Inhibition of GSK3 with lithium treatment still increased caspase-3 activation induced by TRA-8 by approximately threefold in resistant MDA-MB-231 cells. Thus, apoptotic signaling induced by stimulation of TRAIL-R2 was greatly promoted by inhibiting GSK3 with lithium in both susceptible and resistant MDA-MB-231 cells.
Inhibition of GSK3 promotes death receptor-induced caspase-3 activation. (a) MDA-MB-231 parental (left panel) or resistant (right panel) cells were pretreated with 20 mM lithium for 30 min and then treated with the indicated concentrations of TRA-8 for 3 h followed by measurements of caspase-3 activity. (b) 1321N1 cells were pretreated with 20 mM lithium for 30 min and treated with TRAIL-R2 agonistic antibody TRA-8 (1 μg/ml) for the indicated times and caspase-3 activity was measured (left panel) or active caspase-3 and cleaved PARP were measured by immunoblot analysis. (c) MDA-MB-231 cells (left panel) or 1321N1 cells (right panel) were pretreated for 30 min with the indicated GSK3 inhibitors: 20 mM lithium (Li), 5 μM indirubin-3′-monoxime (In), 5 μM kenpaullone (Ke) or 50 μM GSK3 inhibitor 1 (i-1). Cells were then stimulated with TRA-8 and active caspase-3 and cleaved PARP were measured by immunoblot analysis. (d) Cells were pretreated with the indicated GSK3 inhibitors for 30 min, then stimulated with TRAIL-R1 agonistic antibody 2E-12 (2 μg/ml), Fas agonistic antibody CH-11 (10 ng/ml), for SKW6, 4 cells, CH-11 (50 mg/ml for 1321N1 cells), TNFα (50 ng/ml) or TRAIL-R2 agonistic antibody TRA-8 (0.1μg/ml) for the indicated times, and active caspase-3 and cleaved PARP were measured by immunoblot analysis. (e) MDA-MB-231 cells were pretreated with 20 mM lithium for 30 min followed by treatment with the indicated concentrations of TRA-8 or 2E-12 for 16 h and cell death was measured. Quantitative values are means±S.E.M., n=3
Potentiation of TRAIL-R2-mediated activation of caspase-3 by inhibition of GSK3 was also evident in other cell types. In 1321N1 cells, which displayed a very weak apoptosis signal in response to stimulation of TRAIL-R2, GSK3 inhibition with lithium largely increased caspase-3 activation by TRA-8 (Figure 1b). Potentiation of TRAIL-R2-mediated apoptotic signaling by GSK3 inhibition was also evident in immunoblots detecting the TRAIL-R2-mediated production of active cleaved caspase-3 and of caspase-3-induced cleavage product of one of its prominent substrates, poly(ADP-ribose) polymerase (PARP) (Figure 1b). Besides lithium, a panel of structurally diverse selective inhibitors of GSK3 potentiated TRAIL-R2-induced caspase-3 activation in both MDA-MB-231 and 1321N1 cells (Figure 1c). These results demonstrate that endogenous GSK3 provides a strong antiapoptotic regulatory influence on TRAIL-R2-mediated apoptotic signaling.
Inhibition of GSK3 also promoted caspase-3 activation following stimulation of other death receptors (Figure 1d). Stimulation of TRAIL-R1 with the agonistic antibody 2E-12 only slightly activated caspase-3 and caused PARP cleavage in MDA-MB-231 cells, but these responses were greatly increased by treatment with lithium or two other GSK3 inhibitors. Similarly, three GSK3 inhibitors potentiated caspase-3 activation and PARP cleavage following stimulation of Fas in 1321N1 cells. Stimulation of MDA-MB-231 cells with TNFα caused a weak time-dependent activation of caspase-3 and cleavage of PARP, and this was greatly potentiated by three selective GSK3 inhibitors. TRAIL-R2-induced caspase-3 activation was also potentiated by inhibition of GSK3 in two cell lines that undergo apoptosis by type I signaling, SKW6.4 and BJAB cells. The potentiation of TRAIL-R2- and TRAIL-R1-mediated apoptotic signaling by inhibition of GSK3 with lithium was also reflected in measurements of cell death (Figure 1e). These findings that apoptotic signaling is potentiated by a group of structurally diverse GSK3 inhibitors demonstrate that endogenous GSK3 impedes caspase-3 activation induced by stimulation of each of the four major death receptors.
Inhibition of GSK3 promotes death receptor-induced caspase-8 activation and DISC formation
To identify the stage of the apoptosis cascade promoted by inhibiting GSK3, we examined the activation of caspase-8, the initial caspase activated by death receptor stimulation. In MDA-MB-231 cells, inhibition of GSK3 promoted caspase-8 activation as indicated by both measurements of caspase-8 activity and immunoblots of caspase-8 processing following stimulation of TRAIL-R2 (Figure 2a), TRAIL-R1 (Figure 2b) and TNF-R1 (Figure 2c). Inhibition of GSK3 with lithium also promoted Fas- and TRAIL-R2-induced activation of caspase-8 in type I SKW6.4 and BJAB cells (Figure 2d). Thus, the antiapoptotic action of GSK3 is targeted to an early step in apoptosis, the initial activation of caspase-8.
Inhibition of GSK3 promotes death receptor-induced caspase-8 activation and DISC formation. Caspase-8 activity (top) and cleavage of intact procaspase-8 (55/53 kDa) to 43/41 and 18 kDa fragments (bottom) were measured in MDA-MB-231 cells following pretreatment with lithium (20 mM; 30 min) and (a) stimulation of TRAIL-R2 with TRA-8, (b) stimulation of TRAIL-R1 with 2E-12 and (c) stimulation of TNF-R1 with TNFα. Quantitative values are means±S.E.M., n=3. (d) Inhibition of GSK3 with lithium potentiated caspase-8 activation in type I SKW6.4 cells stimulated with agonistic anti-Fas and type I BJAB cells stimulated with TRA-8. (e) MDA-MB-231 cells were treated as indicated with TRA-8 (0.1 μg/ml; 3 h) or lithium (20 mM; 30 min pretreatment) followed by immunoprecipitation of TRAIL-R2 and immunoblotting FADD, caspase-8 and TRAIL-R2. Cell lysates were immunoblotted for caspase-8 to show treatment-dependent cleavage. (f) Jurkat cells were treated as indicated with anti-Fas agonistic antibody (10 ng/ml; 4 h) or lithium (20 mM; 30 min pretreatment) followed by immunoprecipitation of Fas and immunoblotting FADD and caspase-8. Cell lysates were immunoblotted for caspase-8 to show treatment-dependent cleavage
The inhibition of caspase-8 activation by GSK3 suggested that it may impede DISC formation. In MDA-MB-231 cells stimulated with TRA-8 to activate TRAIL-R2, GSK3 inhibition promoted DISC formation as indicated by increased coimmunoprecipitation of FADD and caspase-8 with TRAIL-R2 (Figure 2e), which correlated with increased activation of caspase-8 in cell lysate immunoblots promoted by GSK3 inhibition. Inhibition of GSK3 also enhanced DISC formation following stimulation of Fas in Jurkat cells as indicated by the increased association of FADD and caspase-8 with Fas (Figure 2f), and this also correlated with increased caspase-8 activation in cell lysate immunoblots. Thus, the inhibition of GSK3 promotes death receptor-induced DISC formation, indicating that GSK3 attenuates this initial apoptotic signaling event.
GSK3 associates with the antiapoptotic proteins DDX3 and cIAP-1
We examined if GSK3 associated with other proteins that may be antiapoptotic in the extrinsic apoptotic signaling pathway, specifically DDX3 and cIAP-1, which might allow cooperative actions in controlling apoptosis. These experiments examined only endogenous, not overexpressed, proteins. Immunoprecipitation of GSK3β or GSK3α demonstrated that DDX3 coimmunoprecipitated with each GSK3 isoform from parental and resistant MDA-MB-231 cells, 1321N1, Jurkat and HeLa (Figure 3a).
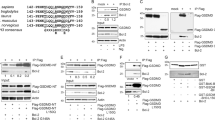
DDX3 is associated with GSK3. (a) DDX3 coimmunoprecipitated with GSK3β or GSK3α from MDA-MB-231 cells, parental (P) or resistant (R), 1321N1 cells, Jurkat cells and HeLa cells. (b) MDA-MB-231 cells were treated with TRA-8 for 0–5 h. Cell lysates were immunoblotted for caspase-3 and DDX3 (FL, full-length protein; CL, cleaved fragment). (c) MDA-MB-231 cells were treated with TRA-8 followed by immunoprecipitation of GSK3β (left panel) or of GSK3α (right panel). Immunoprecipitants were immunoblotted for DDX3 and GSK3. (d) MDA-MB-231 cells, parental or resistant, were treated with TRA-8 for 0–5 h and DDX3 was immunoblotted. (e) MDA-MB-231 cells were pretreated with 15 μM BAF for 1 h and stimulated with TRA-8 for 0, 1, 2, 3 or 4 h, followed by immunoblotting cleaved PARP, intact Bid and intact DDX3. (f) MDA-MB-231 cells were pretreated with 15 μM BAF for 1 h and stimulated with TRA-8 for 0, 3 or 5 h, followed by immunoprecipitation of GSK3β. Immunoprecipitants were immunoblotted for DDX3 and GSK3β. (g) The interaction of GSK3β with domains of DDX3 was studied using the diagramed DDX3 constructs. The cDNAs encoding full-length and truncated DDX3 as indicated were cloned into pcDNA3.1-HisA expression vector. NIH3T3 cells were transfected with the N-terminal-deleted (ND) or C-terminal-deleted (CD) DDX3 constructs and full-length (FL) DDX3, and after 48 h the expressed recombinant DDX3 constructs were detected in cell lysates by immunoblotting using anti-His antibody (right panels). GSK3β was immunoprecipitated and the coimmunoprecipitated recombinant DDX3 was determined by immunoblotting using anti-His antibody (left panels). GSK3β coimmunoprecipitated full-length (FL) DDX3 and the ND2 truncated DDX3, but not the C-terminal-truncated DDX3 constructs
We examined if TRAIL-R2 stimulation affected the association of GSK3β with DDX3, reasoning that it may disrupt the antiapoptotic complex or inactivate one or more proteins to allow apoptosis to proceed. Stimulation of TRAIL-R2 in MDA-MB-231 cells induced activation of caspase-3 time dependently and a concurrent cleavage of DDX3 that led to nearly complete elimination of intact DDX3 (Figure 3b), indicating that the antiapoptotic action of DDX3 is removed by its proteolysis. Coimmunoprecipitation showed that after cleavage a large fragment of DDX3 remained associated with each GSK3 isoform (Figure 3c). There was a notably different effect in resistant MDA-MB-231 cells, as TRAIL-R2 stimulation did not cause cleavage of DDX3 (Figure 3d). To test if DDX3 cleavage following TRAIL-R2 stimulation was caspase-dependent, MDA-MB-231 cells were incubated with the pan-caspase inhibitor BAF. Treatment with BAF blocked the cleavage of two known caspase substrates, PARP and Bid, following TRAIL-R2 stimulation, and also completely blocked DDX3 cleavage (Figure 3e). These results suggest that caspase-dependent cleavage of DDX3 induced by TRAIL-R2 stimulation may relieve the impedance of apoptotic signaling imparted by DDX3.
Considering that full-length DDX3 was depleted after TRAIL-R2 activation, we examined the dynamics of the GSK3β–DDX3 complex. In cells incubated with BAF to block DDX3 cleavage, after TRAIL-R2 stimulation, intact DDX3 remained associated with GSK3β and additional DDX3 was recruited to GSK3β (Figure 3f). Thus, the antiapoptotic proteins, GSK3 and DDX3, are associated in a protein complex, TRAIL-R2 stimulation recruits DDX3 to the GSK3β–DDX3 complex and DDX3 is cleaved to cause depletion of full-length DDX3 by caspases after death receptor activation.
The region of DDX3 that binds to GSK3 was examined by expressing truncated mutants of DDX3 in NIH3T3 cells followed by coimmunoprecipitation (Figure 3g). Truncation of the C-terminal of DDX3, but not its N-terminal, eliminated DDX3 binding to GSK3β, indicating that caspases cleave the N-terminal of DDX3 to allow continued association of a DDX3 fragment containing the C-terminal with GSK3.
Coimmunoprecipitation measurements were also used to test if GSK3 associates with cIAP-1. In untreated MDA-MB-231 cells, cIAP-1 coimmunoprecipitated with GSK3β (Figure 4a). Stimulation of TRAIL-R2 caused time-dependent reductions in GSK3β-associated cIAP-1, with the appearance of a cleaved fragment of cIAP-1 remaining associated with GSK3β. In MDA-MB-231 cells resistant to TRAIL-R2-induced apoptosis, intact cIAP-1 remained associated with GSK3β following stimulation of TRAIL-R2. As with DDX3, blockade of cIAP-1 cleavage with BAF retained intact cIAP-1 associated with GSK3β and caused recruitment of additional cIAP-1 to GSK3β after TRAIL-R2 stimulation (Figure 4b). Thus, GSK3 binds DDX3 and cIAP-1, and these associations are disrupted by cleavage of the GSK3-associated proteins following activation of TRAIL-R2, but they are unaltered by TRAIL-R2 stimulation in apoptosis-resistant cells.
GSK3 associates with cIAP-1. (a) MDA-MB-231 cells, parental or resistant, were treated with TRA-8 for 0–5 h, GSK3β was immunoprecipitated and immunoprecipitants were immunoblotted for cIAP-1 and GSK3β. (b) MDA-MB-231 cells were pretreated with 15 μM BAF for 1 h and stimulated with TRA-8 for 0, 3 or 5 h, followed by immunoprecipitation of GSK3β. Immunoprecipitants were immunoblotted for cIAP-1 and GSK3β
Knockdown of DDX3 with siRNA increases death receptor-induced apoptosis
Experiments knocking down DDX3 were carried out to extend the recent implication that DDX3 inhibits apoptotic signaling stimulated by TRAIL-R2.8 Knockdown of DDX3 by ∼80% accelerated caspase-3 activation caused by stimulation of TRAIL-R2 (Figure 5a), Fas (Figure 5b) or TNF-R1 (Figure 5c). Thus, DDX3 has antiapoptotic actions that slow the rate of caspase-3 activation after death receptor stimulation.
DDX3 inhibits TRAIL-R2-mediated apoptotic signaling. DDX3 levels were reduced using siRNA, and knocked down DDX3 (KD) increased (a) TRA-8-induced caspase-3 activation in Jurkat and HeLa cells, (b) Fas-induced caspase-3 activation in Jurkat cells and (c) TNFα-induced caspase-3 activation in HeLa cells. (d) Knockdown of DDX3 increased caspase-3 activity induced by stimulation of Fas in Jurkat cells or TRAIL-R2 in HeLa cells, and treatment with lithium still potentiated Fas- or TRA-8-induced caspase-3 activation after knockdown of DDX3. Means±S.E.M.; n=3. (e) DDX3 was knocked down in MDA-MB-231 resistant cells, followed by treatment with TRA-8 with or without lithium, and active caspase-3 was immunoblotted
To test if GSK3 and DDX3 act cooperatively to inhibit TRAIL-R2-induced apoptotic signaling, we tested if inhibitors of GSK3 were still capable of promoting apoptosis after knocking down DDX3. DDX3 knockdown increased the activation of caspase-3 following stimulation of Fas or TRAIL-R2 (Figure 5d), and inhibition of GSK3 with lithium still greatly potentiated caspase-3 activation, indicating that DDX3 is not required for the antiapoptotic action of GSK3. Similarly, knockdown of DDX3 in MDA-MB-231 resistant cells increased TRA-8-induced caspase-3 activation, and treatment with lithium greatly increased the activity of caspase-3 (Figure 5e).
TRAIL-R2- and GSK3-associated proteins
As GSK3 acts proximal to the receptor to inhibit DISC formation and GSK3 associates with DDX3 and cIAP-1, we tested if GSK3 and its associated proteins interact with TRAIL-R2, which might signify a receptor-coupled antiapoptotic protein complex. Immunoprecipitation of TRAIL-R2 demonstrated that both GSK3α and GSK3β coimmunoprecipitated (Figure 6a), consistent with GSK3 acting proximal to the receptor.
Stimulation of TRAIL-R2 inhibits GSK3 activity. (a) GSK3α and GSK3β coimmunoprecipitated with TRAIL-R2 in MDA-MB-231 cells. (b) MDA-MB-231 parental (left panel) or resistant (right panel) cells were stimulated with TRA-8, TRAIL-R2 was immunoprecipitated and GSK3 activity was measured in the immunoprecipitants using tau as substrate. Autoradiographs show reduced tau phosphorylation after TRA-8 stimulation of parental cells, but no change in resistant cells. Parallel samples were treated with the GSK3 inhibitor lithium, which blocked GSK3 activity
We tested if TRAIL-R2 stimulation affected TRAIL-R2-associated GSK3 activity. TRAIL-R2 was immunoprecipitated before and after stimulation with TRA-8 and the coimmunoprecipitated GSK3 activity was measured. Stimulation with TRA-8 caused a large decrease (to 35±12% of control; n=5) in the activity of GSK3 that coimmunoprecipitated with TRAIL-R2 (Figure 6b). In contrast, the stimulation of TRAIL-R2 in resistant MDA-MB-231 cells did not inactivate GSK3 (Figure 6b). Thus, GSK3 is associated with TRAIL-R2 and impedes TRAIL-R2-induced apoptotic signaling, and stimulation of TRAIL-R2 overcomes this impedance by inactivating GSK3, but is unable to inactivate GSK3 in resistant cells.
TRAIL-R2-associated proteins were examined in parental and resistant MDA-MB-231 cells. Stimulation of parental, but not resistant, cells caused recruitment of FADD and caspase-8 to TRAIL-R2 and activation of caspase-8 (Figure 7a). In parental MDA-MB-231 cells, DDX3 and cIAP-1 were associated with TRAIL-R2 and were cleaved after stimulation with TRA-8, and the cleaved fragments of each protein remained associated with the receptor (Figure 7b). Cell lysate immunoblots demonstrated a corresponding time-dependent cleavage of total cellular DDX3 and cIAP-1 (Figure 7c). Thus, GSK3, DDX3 and cIAP-1 coexist in a complex and TRAIL-R2 stimulation cleaved DDX3 and cIAP-1 and inactivated GSK3, indicating that TRAIL-R2 activation deactivates the receptor-associated antiapoptotic complex containing GSK3, DDX3 and cIAP-1.
TRAIL-R2-associated proteins. (a) MDA-MB-231 parental and resistant cells were treated with TRA-8 for 0–5 h and TRAIL-R2 immunoprecipitants (left panel) and cell lysates (right panel) were immunoblotted for FADD, caspase-8 and TRAIL-R2. Stimulation of MDA-MB-231 cells, but not resistant cells, caused recruitment to TRAIL-R2 of FADD and caspase-8, and cleavage of caspase-8. (b) MDA-MB-231 cells were treated with TRA-8 for 0–5 h and TRAIL-R2 immunoprecipitants were immunoblotted for DDX3 (FL, full-length protein; CL, cleaved fragment), cIAP-1 and TRAIL-R2 or (c) cell lysates were immunoblotted for DDX3, cIAP-1 and TRAIL-R2. (d) MDA-MB-231 cells were pretreated with 15 μM BAF for 1 h and stimulated with TRA-8 for 0, 3 or 5 h, followed by immunoprecipitation of TRAIL-R2. Immunoprecipitants were immunoblotted for DDX3, cIAP-1, GSK3β and TRAIL-R2. (e) MDA-MB-231 resistant cells were stimulated with TRA-8 for 0–5 h, TRAIL-R2 was immunoprecipitated and immunoprecipitants were immunoblotted for DDX3, cIAP-1 and TRAIL-R2. (f) MDA-MB-231 cells were stimulated with TRAIL-R1 agonistic antibody 2E-12 (2 μg/ml; 24 h) or Jurkat cells were stimulated with Fas agonistic antibody CH-11 (50 ng/ml; 4 h), TRAIL-R1 or Fas was immunoprecipitated and immunoprecipitants were immunoblotted for DDX3, cIAP-1, GSK3β and death receptor. (g) MDA-MB-231 cells were pretreated with 20 mM lithium for 30 min and then stimulated with 50 or 100 ng/ml TRAIL for 4 h followed by immunoblot analysis of cell lysates or immunoprecipitation of TRAIL-R2 and immunoblotting coimmunoprecipitated proteins
The nearly complete cleavage of cellular DDX3 and cIAP-1 and their recruitment to GSK3 after TRAIL-R2 stimulation suggested that DDX3 and cIAP-1 might be recruited to activated TRAIL-R2, perhaps in an attempt to counteract apoptotic signaling. To test this, TRAIL-R2 association with DDX3 and cIAP-1 was assessed after TRAIL-R2 stimulation in the presence of BAF to block caspase-dependent cleavage of DDX3 and cIAP-1. In the presence of BAF, there were increased DDX3 and cIAP-1, above basal levels, associated with TRAIL-R2 after stimulation, indicating that DDX3 and cIAP-1 were recruited to active TRAIL-R2 (Figure 7d). A remarkably similar effect occurred in MDA-MB-231 resistant cells, where little caspase-3 activation occurs after TRAIL-R2 stimulation, but stimulation with TRA-8 caused increased association of DDX3 and cIAP-1 with TRAIL-R2 (Figure 7e). The death receptor association of DDX3, cIAP-1 and GSK3β was also evident in immunoprecipitates of TRAIL-R1 and Fas (Figure 7f), and the stimulation of each receptor induced cleavage of receptor-associated DDX3 and cIAP-1. Interestingly, stimulation of Fas caused recruitment of GSK3β to the receptor.
Receptor-associated complexes were further analyzed after stimulating cells with the endogenous ligand TRAIL that activates both TRAIL-R1 and TRAIL-R2.17 Stimulation of MDA-MB-231 cells with TRAIL led to the activation of caspase-8 and caspase-3 and the corresponding cleavage of PARP, DDX3 and cIAP-1 (Figure 7g). TRAIL-induced caspase activation was increased by GSK3 inhibition with lithium, demonstrating that the same effect is observed with an endogenous death receptor ligand as with agonistic antibodies. Immunoprecipitation of TRAIL-R2 demonstrated the association of GSK3β, DDX3 and cIAP-1 with the receptor before stimulation and the recruitment of FADD and caspase-8 to the receptor after stimulation, as reported earlier.18
Together, these results demonstrate that GSK3, DDX3 and cIAP-1 are associated with TRAIL-R2; DDX3 and cIAP-1 are recruited to TRAIL-R2 upon TRAIL-R2 stimulation; and DDX3 and cIAP-1 are cleaved after receptor stimulation. Knockdown experiments tested if either DDX3 or GSK3 is necessary for the recruitment of this antiapoptotic complex to TRAIL-R2. Knockdown of DDX3 or GSK3β did not change TRAIL-R2 membrane localization, but tended to increase TRAIL-R1 in the membrane (Figure 8a). DDX3 knockdown by ∼80% did not alter the association of GSK3 or cIAP-1 with TRAIL-R2 or cause recruitment of FADD or caspase-8 to the unstimulated receptor (Figure 8b). In contrast, GSK3β knockdown greatly reduced the association of DDX3 and cIAP-1 with TRAIL-R2, but did not cause recruitment of FADD or caspase-8 to the unstimulated receptor (Figure 8c), indicating that GSK3 is necessary for formation of the antiapoptotic complex containing DDX3 and cIAP-1 at TRAIL-R2.
Effects of knocking down GSK3 or DDX3 on TRAIL-R2-associated proteins. (a) Membrane and soluble fractions were prepared from cells with knocked down DDX3 (left panel) or GSK3β (right panel) and immunoblotted for GSK3β, DDX3, TRAIL-R2 and TRAIL-R1. (b) In Jurkat cells, knocking down (KD) the level of DDX3 only modestly reduced the association of cIAP-1 or GSK3β with TRAIL-R2, which were reflected in lower cellular levels of these two proteins after DDX3 knockdown, and did not cause recruitment of FADD or caspase-8 to unstimulated TRAIL-R2. (c) In MDA-MB-231 cells, knocking down the level of GSK3β (3β-KD) almost completely eliminated the association of DDX3 and cIAP-1 with TRAIL-R2, but did not cause recruitment of FADD or caspase-8 to unstimulated TRAIL-R2
Discussion
This study identified an antiapoptotic complex consisting of GSK3, DDX3 and cIAP-1 that caps TRAIL-R2 before stimulation. Inhibiting GSK3 or knocking down DDX3 expression released death receptors from the inhibitory cap to permit more robust apoptotic signaling upon death receptor stimulation. Activation of apoptotic signaling by each of the four major death receptors, TRAIL-R2, TRAIL-R1, Fas or TNF, shares qualitatively similar features, being potentiated by GSK3 inhibition or DDX3 knockdown, indicating that the antiapoptotic protein complex is a common feature of the major receptors inducing extrinsic apoptotic signaling. Extrinsic apoptotic signals overcome this inhibitory cap by disabling the proteins, as stimulation of TRAIL-R2 inactivated GSK3 associated with TRAIL-R2 and induced proteolysis of DDX3 and cIAP-1. In cancer cells resistant to TRAIL-R2-induced apoptosis, GSK3 was not inactivated and DDX3 and cIAP-1 were not cleaved after death receptor stimulation, so they continued to block apoptosis, but inhibiting GSK3 greatly reduced the resistance to death receptor-induced apoptosis. Thus, a complex of GSK3, DDX3 and cIAP-1 provides a checkpoint for initiation of apoptotic signaling by forming a death-antagonizing signaling complex at the death receptor to counterbalance DISC formation and the progression of apoptotic signaling.
GSK3 has a unique role in regulating death receptor-induced apoptosis, being the only enzyme known to act proximal to the four major death receptors to inhibit the initiation of apoptotic signaling by blocking caspase-8 activation. It is worth noting that the greatest apoptosis potentiation by inhibition of GSK3 was achieved with the lowest concentrations of TRA-8 or other conditions where there was only a mild-to-moderate apoptotic signal, suggesting that the checkpoint regulatory mechanism of GSK3 is most effective with relatively weak apoptotic signals and is overcome by stronger stimulation. GSK3 is particularly well suited for this role of maintaining death receptor complexes in an antiapoptotic configuration because GSK3 differs from most kinases by being constitutively active.19, 20 This characteristic of GSK3 not requiring activating signals endows it with the capacity to have a strong influential role in cellular signaling independently of external signals. Moreover, GSK3 is characteristically inactivated by external signals impacting its activity, which differs from most kinases that are activated by receptor-coupled signal transduction mechanisms.15, 21, 22 This GSK3-inactivating mechanism is used by several types of receptors as they generate intracellular signaling cascades. Most widely known is the Wnt pathway, in which plasma membrane receptors generate intracellular signals that by unknown mechanisms inactivate GSK3 complexed with axin, β-catenin and other proteins.23 In addition, growth factor receptors, G protein-coupled receptors, steroid receptors and ionotropic glutamatergic receptors each utilize mechanisms to inactivate GSK3.24, 25, 26, 27 Thus, constitutively active GSK3 is directly or indirectly associated with many types of plasma membrane receptors for which intracellular signaling involves inactivation of GSK3. This study extends to death receptors this characteristic of GSK3, as GSK3 was shown to be associated with TRAIL-R2. In this context, GSK3 counteracts TRAIL-R2-induced apoptotic signaling and, reciprocally, TRAIL-R2 stimulation inactivates GSK3. As with most of these receptor-coupled inhibitory actions of GSK3, the precise mechanism by which GSK3 inhibits receptor-mediated signaling and the mechanism by which stimulation of TRAIL-R2 and several other receptors inactivates GSK3 remain unresolved issues. Although mechanistically our studies focused on the interactions of GSK3 and TRAIL-R2, the results also demonstrate that GSK3 has antiapoptotic actions toward each of the major death receptors and was also associated with TRAIL-R1 and Fas, suggesting that GSK3 likely has similar antiapoptotic mechanisms with each of the death receptors. These results are consistent with the evidence that knocking out GSK3β or inhibiting GSK3 with lithium potentiated TNF-induced apoptosis,13 the first of several studies indicating that GSK3 inhibits extrinsic apoptotic signaling.14, 28, 29, 30 Thus, the association of GSK3 with several classes of membrane receptors and their coupled signaling cascades, as well as the inactivation of GSK3 by receptor stimulation, extends to death receptors. In this way, GSK3 provides a counterbalance to death receptor-induced apoptotic signaling.
In addition to GSK3, antiapoptotic regulation of death receptors was found to be provided by DDX3. Notably, these two antiapoptotic proteins, along with cIAP-1, were found to associate with one another and also with TRAIL-R2. We tested if DDX3 scaffolds GSK3 or cIAP-1 to TRAIL-R2, but knocking down DDX3 did not alter their associations with TRAIL-R2. On the other hand, knocking down GSK3 reduced the association of DDX3 and cIAP-1 with TRAIL-R2. Thus, GSK3 is critical for the formation of the death-antagonizing signaling complex at the receptor. Blocking the action of DDX3 by knocking down its levels promoted death receptor-induced signaling, but the mechanism for this action of DDX3 remains to be identified. Furthermore, death receptor activation caused rapid cleavage of DDX3, presumably to inactivate its antiapoptotic action.7 Examining the mechanism by which DDX3 protects from apoptotic signaling is hampered by the limited information available about DDX3 functions. DDX3 is a DEAD box protein family member, named for the conserved D-E-A-D sequence and putative RNA helicase actions,31, 32 but few functions of DDX3 have been identified, and these generally involve mRNA.33, 34, 35 However, other DEAD box proteins have a variety of actions apparently unrelated to modulation of mRNA.36 Interestingly, one prominent DEAD box protein, p68/DDX5, interacts with the β-catenin–GSK3 signaling complex,37 raising the possibility that DEAD box proteins have multiple interactions with GSK3-associated signaling events. The question remains of how the antiapoptotic actions of GSK3 and DDX3 are coordinated. Each is associated with TRAIL-R2, and the inhibition of either protein promotes TRAIL-R2-induced apoptotic signaling. It is possible that their association facilitates their antiapoptotic actions or that the presence of two antiapoptotic proteins provides redundancy for inhibiting apoptotic signaling.
In summary, death receptors were found to be capped by an antiapoptotic protein complex containing GSK3, DDX3 and cIAP-1. Stimulation of death receptors disables these proteins, causing inactivation of GSK3 and cleavage of DDX3 and cIAP-1 to permit progression of the apoptotic signal. Impairment of the death receptor-induced disabling of these proteins may contribute to resistance to death receptor-induced apoptosis in some cancer cells, suggesting that targeting GSK3 and DDX3 may be a useful strategy for promoting death receptor-induced apoptosis.
Materials and Methods
Cell culture and materials
HeLa cells and MDA-MB-231 human breast cancer cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Cellgro, Herndon, VA, USA) in humidified 37 °C chambers with 5% CO2. Human astrocytoma 1321N1 cells were grown in DMEM supplemented with 5% fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin. Jurkat, SKW6.4 and BJAB cells were grown in RPMI-1640 medium (Cellgro) supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 2.5 g/l glucose, 100 U/ml penicillin and 100 μg/ml streptomycin. Sources of chemicals were lithium chloride (Sigma-Aldrich, St. Louis, MO, USA), indirubin-3′-monoxime, GSK3 inhibitor 1, kenpaullone, TNFα (CalBiochem, San Diego, CA, USA), human recombinant TRAIL and pan-caspase inhibitor Boc-D(OMe)-FMK (BAF; Alexis Biochemicals, San Diego, CA, USA). Sources of antibodies were caspase-3, cleaved active caspase-3, cleaved 85 kDa fragment of PARP, caspase-8 for cell lysate immunoblots, FADD (Cell Signaling Technology, Beverly, MA, USA), caspase-8 for DISC analysis (Immunotech, Marseille, France), TRAIL-R1 for western blots (BD Pharmingen, San Diego, CA, USA), β-actin (Sigma), Fas antibody CH-11 (Transduction Laboratories, Lexington, KY, USA), TRAIL-R2 antibody for western blots (Prosci Inc., Poway, CA, USA), Fas antibody for western blots (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-His-tag antibody (Upstate Biotechnology Inc., Lake Placid, NY, USA). Monoclonal antibodies TRA-8 to stimulate TRAIL-R2 and 2E-12 to stimulate TRAIL-R1, monoclonal TRAIL-R2 antibody 2B9 for immunoprecipitation, monoclonal TRAIL-R1 antibody 2B4 for immunoprecipitation and antibodies to DDX3 and cIAP-1 were generated in the laboratory of Dr. T Zhou.8 Horseradish peroxidase-conjugated GSK3α/β and all horseradish peroxidase-conjugated secondary antibodies were purchased from Southern Biotechnology Associates (Birmingham, AL, USA). siRNA constructs were from Sigma-Aldrich and used according to the manufacturer's protocols. Cell death was determined using the Live/Dead kit according to the manufacturer's instructions (Invitrogen). Membrane fractions were prepared using the Native Membrane Protein Extraction Kit (Calbiochem) according to the manufacturer's protocol.
Immunoblotting and immunoprecipitation
After washing with PBS twice, cells were harvested in lysis buffer (20 mM Tris (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1 mM sodium orthovanadate, 100 μM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 5 μg/ml pepstatin, 50 mM NaF, 1 nM okadaic acid, 1% Triton X-100 and 10% glycerol), centrifuged at 20 800 × g for 15 min, and supernatants were collected. Protein concentrations were determined using the bicinchoninic method (Pierce, Rockford, IL, USA). Cell lysates were mixed with Laemmli sample buffer and placed in a boiling water bath for 5 min. Proteins (10–20 μg) were resolved in SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were probed with the indicated primary antibodies, and immunoblots were developed using horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit IgG, followed by detection with enhanced chemiluminescence.
For immunoprecipitation of TRAIL-R2 or TRAIL-R1, cell lysates (1 mg protein) were incubated with 40 μl of sepharose 4B-conjugated anti-TRAIL-R2 monoclonal antibody 2B9 (which does not compete with TRA-8 for binding TRAIL-R2) or anti-TRAIL-R1 monoclonal antibody 2B4 (which does not compete with 2E-12 for binding TRAIL-R1) for 2 h at 4 °C. For immunoprecipitation of GSK3β or GSK3α, cell lysates (500 μg) were incubated with 20 μl of sepharose 4B-conjugated anti-GSK3β monoclonal antibody or anti-GSK3α monoclonal antibody (Southern Biotech) for 2 h at 4 °C. The samples were then washed three times with 400 μl lysis buffer, suspended in 40 μl of Laemmli buffer, placed in a boiling water bath for 5 min and the proteins resolved by electrophoresis. For immunoprecipitation of Fas, cell lysates (1 mg protein) were precleared by incubating with protein G for 30 min at 4 °C, centrifuged, the supernatants were incubated with 1 μg anti-Fas antibody CH-11 overnight, 5 μg goat anti-mouse IgM antibody was added and the samples shaken for 2 h at 4 °C. The samples were then incubated with 30 μl of protein G sepharose beads for 2 h, placed in a boiling water bath for 5 min and the proteins resolved by electrophoresis. For immunoblotting after immunoprecipitation, antibodies used were anti-TRAIL-R2 from Prosci Inc., anti-TRAIL-R1 from BD Pharmingen, anti-Fas from Santa Cruz, anti-caspase-8 from Immunotech and anti-FADD from Cell Signaling.
Caspase activities
Fluorometric assays of caspase-3 or caspase-8 activity were conducted as described earlier38 in 96-well clear-bottom plates, and all measurements were carried out in triplicate wells. To each well, 200 μl of assay buffer (20 mM HEPES (pH 7.5), 10% glycerol, 2 mM dithiothreitol) was added. The peptide substrate for caspase-3 DEVD-AMC or caspase-8 IETD-AMC (Alexis Biochemicals) was added to each well to a final concentration of 25 ng/μl. Cell lysates (20 μg protein) were added to start the reaction. Fluorescence was measured on a fluorescence plate reader (Bio-Tek, Winooski, VT, USA) set at 360 nm excitation and 460 nm emission. Caspase activity was calculated as ((mean AMC fluorescence from triplicate wells) – (background fluorescence ))/μg protein.
GSK3 activity measurements
GSK3 activity was measured after immunoprecipitation of TRAIL-R2 using either a primed phosphorylated peptide substrate or using recombinant tau as substrate, as described earlier,39 both of which gave equivalent results. TRAIL-R2 was immunoprecipitated and the beads washed twice in PBS/BSA and once in kinase buffer (20 mM Tris (pH 7.5), 5 mM MgCl2 and 1 mM dithiothreitol). The beads were then resuspended in 30 μl kinase buffer containing 250 μM ATP, 1.4 μCi [γ-32P]-ATP and 50 μM phospho-glycogen synthase peptide (Upstate biotechnology Inc.). The samples were incubated at 30 °C for 30 min, the reaction tubes were centrifuged for 1 min and triplicate 9-μl aliquots of each sample were spotted on P81 filter paper circles. The filter paper was allowed to dry and then washed three times in 0.5% phosphoric acid for a total time of 1 h, followed by a 10-min wash in 95% ethanol. The filter paper was then air-dried and counted in a liquid scintillation counter. Alternatively, the kinase reaction was carried out after TRAIL-R2 immunoprecipitation with 0.1 μg/μl recombinant tau protein (Panvera, Madison, WI, USA) for 30 min at 30 °C, and 25 μl of Laemmli sample buffer (2% SDS) was added to stop the reaction. Samples were placed in a boiling water bath for 5 min, proteins were separated in 8% SDS polyacrylamide gels, gels were vacuum-dried, exposed to a phosphoscreen overnight and quantitated using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA).
Abbreviations
- cIAP-1:
-
cellular inhibitor of apoptosis protein-1
- DISC:
-
death-inducing signaling complex
- FADD:
-
Fas associated with death domain protein
- GSK3:
-
glycogen synthase kinase-3
- TNF:
-
tumor necrosis factor
- TRAIL-R1/2:
-
TNF-related apoptosis-inducing ligand receptor-1/2
References
Ashkenazi A, Dixit VM . Death receptors: signaling and modulation. Science 1998; 281: 1305–1308.
Peter ME, Krammer PH . The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ 2003; 10: 26–35.
Kaufmann SH, Hengartner MO . Programmed cell death: alive and well in the new millennium. Trends Cell Biol 2001; 11: 526–534.
Dutton A, Young LS, Murray PG . The role of cellular FLICE inhibitory protein (c-FLIP) in the pathogenesis and treatment of cancer. Expert Opin Ther Targets 2006; 10: 27–35.
Rothe M, Pan MG, Henzel WJ, Ayres TM, Goeddel DV . The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 1995; 83: 1243–1252.
Shu HB, Takeuchi M, Goeddel DV . The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci USA 1996; 93: 13973–13978.
Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin Jr AS . NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998; 281: 1680–1683.
Li Y, Wang H, Wang Z, Makhija S, Buchsbaum D, Lobuglio A et al. Inducible resistance of tumor cells to tumor necrosis factor-related apoptosis-inducing ligand receptor 2-mediated apoptosis by generation of a blockade at the death domain function. Cancer Res 2006; 66: 8520–8528.
Beurel E, Jope RS . The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog Neurobiol 2006; 79: 173–189.
Beyaert R, Vanhaesebroeck B, Suffys P, Van Roy F, Fiers W . Lithium chloride potentiates tumor necrosis factor-mediated cytotoxicity in vitro and in vivo. Proc Natl Acad Sci USA 1989; 86: 9494–9498.
Beyaert R, Heyninck K, De Valck D, Boeykens F, van Roy F, Fiers W . Enhancement of tumor necrosis factor cytotoxicity by lithium chloride is associated with increased inositol phosphate accumulation. J Immunol 1993; 151: 291–300.
Klein PS, Melton DA . A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA 1996; 93: 8455–8459.
Hoeflich KP, Luo J, Rubie EA, Tsao M-S, Jin O, Woodgett JR . Requirement for glycogen synthase kinase-3β in cell survival and NF-κB activation. Nature 2000; 406: 86–90.
Schwabe RF, Brenner DA . Role of glycogen synthase kinase-3 in TNF-α-induced NF-κB activation and apoptosis in hepatocytes. Am J Physiol Gastrointest Liver Physiol 2002; 283: G204–G211.
Manoukian AS, Woodgett JR . Role of glycogen synthase kinase-3 in cancer: regulation by Wnts and other signaling pathways. Adv Cancer Res 2002; 84: 203–229.
Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med 2001; 7: 954–960.
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995; 3: 673–682.
Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A . Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity 2000; 12: 611–620.
Frame S, Cohen P . GSK3 takes centre stage more than 20 years after its discovery. Biochem J 2001; 359: 1–16.
Jope RS, Johnson GV . The glamour and gloom of glycogen synthase kinase-3. Trends Biochem Sci 2004; 29: 95–102.
Grimes CA, Jope RS . The multi-faceted roles of glycogen synthase kinase-3β in cellular signaling. Prog Neurobiol 2001; 65: 391–426.
Woodgett JR . Physiological roles of glycogen synthase kinase-3: potential as a therapeutic target for diabetes and other disorders. Curr Drug Targets Immune Endocr Metabol Disord 2003; 3: 281–290.
Ciani L, Salinas PC . WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci 2005; 6: 351–362.
Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA . Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995; 378: 785–789.
Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR et al. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc Natl Acad Sci USA 2004; 101: 5099–5104.
Cardona-Gomez P, Perez M, Avila J, Garcia-Segura LM, Wandosell F . Estradiol inhibits GSK3 and regulates interaction of estrogen receptors, GSK3, and β-catenin in the hippocampus. Mol Cell Neurosci 2004; 25: 363–373.
Peineau S, Taghibiglou C, Bradley C, Wong TP, Liu L, Lu J et al. LTP inhibits LTD in the hippocampus via regulation of GSK3β. Neuron 2007; 53: 703–717.
Liao X, Zhang L, Thrasher JB, Du J, Li B . Glycogen synthase kinase-3β suppression eliminates tumor necrosis factor-related apoptosis-inducing ligand resistance in prostate cancer. Mol Cancer Ther 2003; 2: 1215–1222.
Song L, Zhou T, Jope RS . Lithium facilitates apoptotic signaling induced by activation of the Fas death domain-containing receptor. BMC Neurosci 2004; 5: 20–27.
Rottmann S, Wang Y, Nasoff M, Deveraux QL, Quon KC . A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3β/FBW7 loss of function. Proc Natl Acad Sci USA 2005; 102: 15195–15200.
Abdelhaleem M, Maltais L, Wain H . The human DDX and DHX gene families of putative RNA helicases. Genomics 2003; 81: 618–622.
Rocak S, Linder P . DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 2004; 5: 232–241.
Kanai Y, Dohmae N, Hirokawa N . Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 2004; 43: 513–525.
Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 2004; 14: 685–691.
Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT . Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004; 119: 381–392.
Fuller-Pace FV . DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 2006; 34: 4206–4215.
Yang L, Lin C, Liu ZR . P68 RNA helicase mediates PDGF-induced epithelial mesenchymal transition by displacing Axin from β-catenin. Cell 2006; 127: 139–155.
Bijur GN, De Sarno P, Jope RS . Glycogen synthase kinase-3β facilitates staurosporine- and heat shock-induced apoptosis: Protection by lithium. J Biol Chem 2000; 275: 7583–7590.
Watcharasit P, Bijur GN, Zmijewski JW, Song L, Zmijewska A, Chen X et al. Direct, activating interaction between glycogen synthase kinase-3β and p53 after DNA damage. Proc Natl Acad Sci USA 2002; 99: 7951–7955.
Acknowledgements
This research was supported by grants AG021045, MH38752 and CA112169 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by J Tschoop
Rights and permissions
About this article
Cite this article
Sun, M., Song, L., Li, Y. et al. Identification of an antiapoptotic protein complex at death receptors. Cell Death Differ 15, 1887–1900 (2008). https://doi.org/10.1038/cdd.2008.124
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.124
Keywords
This article is cited by
-
Molecular docking, synthesis, and biological evaluation of 7-azaindole-derivative (7AID) as novel anti-cancer agent and potent DDX3 inhibitor:—an in silico and in vitro approach
Medical Oncology (2022)
-
DDX3X: structure, physiologic functions and cancer
Molecular Cancer (2021)
-
The DEAD-box protein family of RNA helicases: sentinels for a myriad of cellular functions with emerging roles in tumorigenesis
International Journal of Clinical Oncology (2021)
-
Tamoxifen induction of Cre recombinase does not cause long-lasting or sexually divergent responses in the CNS epigenome or transcriptome: implications for the design of aging studies
GeroScience (2019)
-
GSK3 suppression upregulates β-catenin and c-Myc to abrogate KRas-dependent tumors
Nature Communications (2018)