Abstract
The outcome of a viral infection depends on the interplay between the host's capacity to trigger potent antiviral responses and viral mechanisms that counteract them. Although Toll-like receptor (TLR)-3, which recognizes virally derived double-stranded (ds) RNA, transmits downstream antiviral signaling through the TIR adaptor Trif (TICAM-1), viral RNA-sensing RIG-like helicases (RLHs) use the mitochondrial-bound CARD protein Cardif (IPS-1/MAVS/VISA). The importance of these two antiviral signaling pathways is reflected by the fact that both adaptors are inhibited through specific cleavage triggered by the hepatitis C virus serine protease NS3-4A. Here, we show that inactivation can also occur through cellular caspases activated by various pro-apoptotic signals. Upon caspase-dependent cleavage both adaptors loose their capacity to activate the transcription factors interferon regulatory factors (IRF) and NF-κB. Importantly, poliovirus infection triggers a caspase-dependent cleavage of Cardif, suggesting that some viruses may activate caspases not only as a mean to facilitate shedding and replication, but also to impair antiviral responses.
Similar content being viewed by others
Main
During microbial infection, it is crucial for the host to rapidly mount efficient innate immune responses. Several pathogen recognition systems exist to ensure such an innate response. The best-known examples are the transmembrane Toll-like receptors (TLR), which in most cases recognize extracellular pathogen-associated molecular patterns. These pathogen-associated molecular patterns are essential molecules for the microbes and are diverse in composition, including sugars, lipids, proteins and nucleic acids.1, 2 All TLRs initiate intracytoplasmic signaling events through their Toll-interleukin-1 receptor (TIR) domain by recruiting TIR domain-containing adaptor molecules through homotypic (TIR–TIR) interactions. Five TIR adaptors have been described so far in mammals.3 One of them, MyD88, is recruited to all TLRs, with the exception of TLR3. Upon activation by double-stranded (ds) RNA, TLR3 triggers downstream signaling through TIR domain-containing adapter inducing IFN-β (Trif), another TIR adaptor.4, 5, 6
TLR3 serves antiviral functions through the sensing of virally derived dsRNA species within endosomal compartments. TLR7/8 and TLR9 are other endosome-associated nucleic acid sensors, which detect single-stranded (ss)RNA and non-methylated CpG DNA, respectively. All these TLRs, when detecting foreign nucleic acids within endosomes, activate the transcription factors interferon regulatory factors (IRF), IRF3 and IRF7, which trigger the production of antiviral type-I interferons (IFN).7 More recently, other, TLR-independent, antiviral surveillance systems have been described, which rely on the detection of cytoplasmic RNA of viral origin by RIG-like helicases. Two of them, RIG-I and MDA5, have been well characterized.8, 9 Although both helicases use their N-terminal caspase recruitment domains (CARD) to recruit the mitochondrial-bound CARD adaptor Cardif (CARD adaptor-inducing interferon-β; also called IPS-1, MAVS and VISA), they sense different viruses.10, 11, 12, 13, 14, 15 In-depth analysis of the corresponding knockout mice revealed that MDA5 mounts innate responses to picornaviruses, whereas RIG-I is activated by certain flaviviruses, vesicular stomatitis virus (VSV), Sendai virus and influenza virus.16, 17
The importance of antiviral systems is in parallel with the number of viruses that target the signaling pathway. In support of this notion, both Trif and Cardif are specifically cleaved and inactivated by the hepatitis C virus (HCV) NS3-4A serine protease.12, 18 Although NS3-4A-mediated cleavage induces the release of Cardif from the mitochondria, thereby impairing downstream signaling events,19, 20 cleavage of Trif is believed to preclude recruitment and assembly of effector proteins. Hepatitis A virus (HAV) also cleaves and inactivates Cardif, further supporting its essential role in antiviral defense.21
The importance of Cardif and Trif in the antiviral response prompted us to postulate that other viruses might also target these effectors. In principle, viruses may use their own proteases (such as HCV and HAV) or, alternatively, activate cellular proteases, which in turn cleave the cellular proteins. As viral infection is frequently associated with cell death, we reasoned that caspases might represent potential proteases for Cardif or Trif cleavage. Here, we show that both adaptors are efficiently cleaved by caspases, leading to their inactivation. Importantly, caspase activation initiated during infection with poliovirus causes Cardif cleavage. As poliovirus belongs to the picornavirus family that is sensed by MDA5, poliovirus may target Cardif for cleavage as a mean to evade innate immune responses.
Results
Cardif and Trif are cleaved by caspases
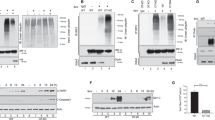
To determine whether caspases could cleave Cardif, we triggered apoptosis in a U-2 OS-derived cell line by adding recombinant Fas ligand (FasL) (Figure 1a). A Cardif fragment of approximately 50 kDa was generated, which was smaller in size than the 70 kDa fragment generated by the HCV NS3-4A serine protease (Figure 1a).12, 19, 22, 23 As expected, FasL, but not NS3-4A-mediated cleavage, was caspase-dependent, because incubation with the broad caspase inhibitor zVAD only blocked the former (Figure 1a).
Cardif is cleaved by caspases. (a) U-2 OS human osteosarcoma-derived cell lines inducibly expressing NS3-4A upon tetracycline (Tet) withdrawal were treated with 100 ng/ml of Fc:FasL for 6 h with or without zVAD (25 μM). Where indicated NS3-4A was induced by the removal of Tet 18 h before Fc:FasL treatment. Cell extracts were analyzed by immunoblotting. (b) Jurkat T cells, either wild type (WT) or caspase-8 deficient, were stimulated with the indicated pro-apoptotic stimuli for 6 h in the presence or absence of zVAD. Cell extracts were analyzed by immunoblotting. (c) HEK293T-6 cells were transfected with the indicated VSV-Cardif constructs. zVAD was added after the removal of the transfection medium (8 h after transfection). At 18 h after transfection, cells were treated with 100 ng/ml Fc:FasL for 6 h and cell extracts were analyzed by immunoblot. (d) Structural organization of Cardif. CARD, caspase recruitment domain; TR, Traf3 binding site; TM, transmembrane domain
Stimulation of cells with other apoptosis inducers, such as staurosporine, or the DNA-damaging agents etoposide, doxorubicin and camptothecin also resulted in Cardif cleavage in a Jurkat T-cell line (Figure 1b), demonstrating that the triggering of various pro-apoptotic pathways can result in Cardif processing. Recently, Cardif was shown to interact with caspase-8, most likely indirectly via Fas-associated death domain protein (FADD).24 We therefore tested the possibility that this caspase was responsible for Cardif cleavage by using a Jurkat T-cell clone that lacks caspase-8.25 However, in this setting, Cardif cleavage still occurred upon incubation with the different apoptosis inducers, with the exception of FasL as expected (caspase-8 being a crucial component of the Fas signaling pathway) (Figure 1b). This strongly suggests that caspase-8 does not cleave Cardif directly. Notably, Cardif was also cleaved upon FasL stimulation in caspase-9-deficient Jurkat T cells (Supplementary Figure S1). Hence, in an effort to identify further which caspase(s) was responsible for Cardif cleavage, we used commercially available inhibitors to caspase-3, -6, -8, -9 and -10. However, all inhibitors interfered with Cardif cleavage to some extent (data not shown), most likely owing to a lack of specificity, as reported recently.26 Furthermore, caspase-3 knockdown Jurkat T cells still showed Cardif and Trif cleavage upon FasL and etoposide stimulation, suggesting either that other effector caspases, such as caspase-6 or -7, cleave these adaptors or that caspase-3 knockdown was not sufficient to observe diminished cleavage (Supplementary Figure S2).
We then looked for a putative caspase cleavage site and found one interesting candidate, D429, in an SQVD motif of the Cardif gene. This sequence is highly similar to the SQLD caspase-targeted sequence found in the kinase RIP4, an activator of NF-κB.27 We mutated D429 into E and overexpressed wild-type (WT), Cardif C508A (a point mutant that is resistant to NS3-4A-mediated cleavage)12 and Cardif D429E mutant in 293T-6 cells (Figure 1c and d). 293T-6 cells, which originate from 293T, were used for their higher sensitivity to FasL as compared with the parental cells.28 The D429E mutant construct, but neither WT nor C508A Cardif, was resistant to FasL-induced cleavage (Figure 1c), demonstrating that Cardif is indeed cleaved by caspases at D429. When the cleaved fragment was purified from a Coomassie gel and analyzed by mass spectrometry after tryptic digestion, a non-tryptic peptide ending at amino acid D429 was obtained, which further supports the Cardif SQVD motif as the cleavage site targeted by caspases (data not shown). Hence, Cardif is cleaved near the C terminus by a caspase during apoptosis triggered by different stimuli.
Upon HCV infection, the TLR3 adaptor molecule Trif is cleaved by HCV NS3-4A, which is similar to the Cardif protein.18 In addition, triggering a Trif-dependent pathway can result in apoptosis, most likely because Trif interacts with RIP1, which, through its death domain, recruits FADD that in turn binds to caspase-8.29, 30, 31, 32 Collectively, this knowledge prompted us to test whether Trif was also subjected to cleavage by caspases. When overexpressed, Trif potently induced apoptosis, as inferred by cellular morphology and PARP cleavage (Figure 2a and data not shown), and was efficiently cleaved in a caspase-dependent manner (Figure 2a). When scrutinizing for putative caspase sequences within the Trif molecule, we found the three aspartate residues, D281 (VAPD), D289 (GLPD) and D340 (SVED), to be the most likely caspase targets (Figure 2b). Therefore, we generated point mutations at the corresponding amino acid positions. When N terminally VSV-tagged WT, D289E and D340E were expressed, a cleaved fragment of approximately 34 kDa appeared in the absence of zVAD (Figure 2a). This cleaved fragment most likely corresponds to amino acids 1–281. In contrast, when the D281E point mutant was expressed, another cleaved fragment, approximately 1 kDa larger, appeared. This suggested that another aspartate residue, localized about 10 amino acids after D281, was also the target of cleavage by caspases. Indeed, a construct in which both D281 and D289 were mutated to Glu (Trif DDEE) was resistant to cleavage. Hence, both D281 and D289 are targeted by the activity of caspases. Importantly, the endogenous Trif molecule was also cleaved in response to FasL or etoposide stimulation in Jurkat T cells (Figure 2c). In that case, although Trif cleavage was abrogated in FasL-stimulated caspase-8-deficient cells, it still occurred upon etoposide treatment, suggesting that caspase-8 does not directly cleave Trif. Hence, similar to Cardif, the Trif adaptor is processed not only by HCV NS3-4A but also by caspases.
Trif is cleaved by caspases. (a) 293T-6 cells were transfected with the indicated VSV-Trif constructs. Where indicated zVAD was added after the removal of the transfection medium. At 18 h after transfection, cells were treated with 100 ng/ml of Fc:FasL for 6 h and cell extracts were analyzed by immunoblotting. (b) Structural organization of TRIF. R, RIP homotypic interaction motif (RHIM); TIR, Toll-interleukin-1 receptor. (c) Jurkat T cells were stimulated with 100 ng/ml of Fc:FasL or etoposide (at the indicated concentrations) for 6 h in the presence or absence of zVAD. Anti-Trif immunoprecipitates (IP) and cell extracts (XT) were analyzed by immunoblot. *Ig(H), immunoglobulin heavy chain
Cleavage of Cardif and Trif results in their inactivation
To test the functional consequences of caspase-mediated cleavage, we generated for each adaptor two deletion constructs representing the N- and C-terminal cleavage fragments, that is Cardif 1–429, Cardif 430–540, Trif 1–281 and Trif 290–712, respectively (Figures 1d and 2b). When individually expressed in cells with an IFN-β or NF-κB-dependent promoter, we observed that none of the constructs activated the reporter gene (Figure 3a). Regarding Cardif, these results are in agreement with previous findings that showed that when either the C-terminal transmembrane or the N-terminal caspase recruitment domains is removed, the activity of the molecule is abolished.10, 12, 13, 15 The Trif C-terminal construct (290–712) was the only exception retaining NF-κB activation potential (Figure 3b), most likely because of an ongoing interaction with RIP1 that results in NF-κB activation.31 In contrast, the IRF-responsive ISRE Luciferase reporter gene was not activated by any of the deletion mutants (Figure 3c), whereas both Cardif and Trif caspase-resistant point mutant constructs were as efficient as WT counterparts at promoting IFN-β, NF-κB and ISRE reporter activations (Figure 3a–c). Moreover, we noticed no inherent differences between Cardif D429E and WT constructs at promoting antiviral responses, as both molecules showed comparable antiviral activities when introduced into Cardif knockdown cells (Supplementary Figure S3). Interestingly, upon TLR3 triggering, Trif cleavage occurred concomitantly to diminished NF-κB and IRF3 activations (Supplementary Figure S4), suggesting that caspase-mediated cleavage might serve to dampen or limit innate responses over time.
Cardif- and Trif-mediated IRF and NF-κB activation are compromised by caspase cleavage. 293T cells were transfected with (a) an IFN-β, (b) an NF-κB or (c) an IRF-responsive (ISRE) reporter plasmid together with an empty plasmid or the indicated Cardif (left) or Trif (right) constructs (at two different doses in panels b and c) and analyzed for luciferase activity. Data are the mean values from one experiment representative of three independent experiments, each performed in triplicates. Error bars represent the SD of triplicates of the experiment presented
We next evaluated the endogenous levels of proteins regulated by Cardif and Trif activities. As the expression of both RIG-I and MDA5 is induced during antiviral responses through autocrine and paracrine IFN signaling,9, 33 we monitored Cardif- and Trif-mediated antiviral activities by analyzing the expressions of the two helicases. Both RIG-I and MDA5 protein levels were induced by WT and point mutant Cardif and Trif constructs, whereas no expression could be detected upon transfection of the deletion mutants or an empty vector (Figure 4a). Furthermore, when monitored by ELISA, no or only marginal production of RANTES was detected upon expression of any of the four deletion constructs in marked contrast to WT and point mutant forms of Cardif and Trif (Figure 4b). Altogether, these results indicate that the promotion of antiviral responses is largely compromised when Cardif and Trif are cleaved by caspases, with the C-terminal Trif cleavage product retaining NF-κB activation potential.
Cardif- and Trif-mediated protein induction is impaired by caspase cleavage. (a) 293T cells were transfected with an empty plasmid or the indicated VSV or Flag Cardif (left) or Trif (right) constructs, as indicated and analyzed for RIG-I and MDA5 expression by immunoblotting. *Nonspecific bands. (b) 293T cells were transfected with an empty plasmid or the indicated Cardif (left panel) or Trif (right panel) constructs. At 24 h after transfection, cell supernatants were analyzed for RANTES expression by ELISA. Data are the mean values from one experiment representative of two independent experiments, each performed in triplicates. Error bars represent the SD of triplicates of the experiment presented
Poliovirus infection results in caspase-dependent Cardif cleavage
Next, we tested whether the cleavage of Cardif by caspases may be relevant in the context of a viral infection. As picornaviruses such as encephalomyocarditis virus are recognized by MDA5,16, 17 and, more recently, the polio picornavirus was reported to cleave MDA5 in a caspase-dependent manner,34 we sought to determine whether poliovirus also uses caspases to cleave Cardif. In HeLa cells, Cardif cleavage became apparent 8 h after poliovirus infection (Figure 5). Although HAV, another picornavirus family member, was recently reported to directly cleave Cardif through its 3ABC protease, it is unlikely that poliovirus proteases cleave Cardif. Indeed, in poliovirus-infected cells, cleavage of Cardif was completely blocked by zVAD (Figure 5), suggesting that poliovirus-induced caspase activation is responsible for Cardif cleavage.
Poliovirus infection results in caspase-dependent Cardif cleavage. HeLa cells were infected for the indicated time periods with poliovirus at a multiplicity of infection of 5. zVAD was added for the same time periods when indicated. Cells were harvested, lysed in RIPA buffer and analyzed for the cleavage of Cardif by immunoblotting. *Nonspecific bands
Discussion
During recent years, many efforts have been made toward elucidating the pathways implicated in the host defense against viruses. Crucial molecules were discovered, and novel pathways were identified and characterized. RIG-like helicases and TLR3 signaling offer a remarkable example of such newly described pathways, conferring to the host a potent antiviral immunity. By interfering with type-I IFN activities mediated by TLR3 and RIG-like helicases, viruses gain survival advantages. This strategy is indeed used by many viruses, including paramyxoviruses whose V protein directly binds MDA5.8 Cardif is cleaved by viral proteases present in HAV and HCV.12, 21 In this report, we unravel one additional strategy, which can be used by viruses to dampen the activity of these two pathways. We found that both the adaptor molecules Cardif and Trif undergo caspase-dependent cleavage and inactivation during apoptosis.
Although at present it is still unclear which caspase(s) is responsible for their cleavage, it is unlikely to be caspase-9 or -8, although the latter has been reported to be indirectly recruited to both Cardif and Trif, probably through FADD.24, 30, 32 Indeed, our data show that Cardif and Trif are cleaved by several pro-apoptotic stimuli in caspase-8 (and caspase-9)-deficient cells, and also in caspase-3 knockdown cells. Interestingly, different caspases might serve distinct functions in antiviral responses. For example, caspase-8 was shown to be essential for Cardif-induced NF-κB activation,24 whereas the caspase-8 adaptor FADD mediates an antiviral response against VSV.35 Here, we demonstrate that other (non-caspase-8) activities antagonize Cardif and Trif functions. These apparently opposing roles might be reconciled when considering the duration of an antiviral response. Indeed, in a first step (the innate defense step), TLR3 and RIG-I, sensing the presence of viruses, recruit caspase-8 via Cardif and Trif to promote inflammatory and antiviral reactions through NF-κB and IRF activation. In a later step, the virus counter-attacks by activating additional caspases that in turn inactivate the two antiviral pathways through cleavage of Cardif and Trif. In some cases such as poliovirus, the caspase activity might be high enough to trigger apoptosis. A second model is that caspase cleavage of Trif and Cardif represents a host mechanism to regulate the extent and/or duration of antiviral responses. This hypothesis is supported by the observation of Trif cleavage upon TLR3 triggering by poly(I:C) (Supplementary Figure S4). Notably, the Trif C-terminal cleavage fragment still retains some NF-κB activation potential, suggesting that it might form a cytoplasmic complex with RIP1, allowing NF-κB responses to persist during cell death. In contrast, Cardif activities are completely blocked by caspase-mediated cleavage, which could constitute a general mechanism dampening inflammatory reactions during cell death (third hypothesis).
Although many viruses actively inhibit apoptosis, there are also others that activate it.36 Some viruses may induce apoptosis as a mean for dissemination, and therefore, apoptosis may be an important step in the viral life cycle.37, 38 It is also now recognized that some viruses directly use components of the apoptotic pathway to facilitate their replication.38 The nonstructural protein from the B19 parvovirus induces apoptosis in various cell lines. Infection by Sindbis virus results in apoptosis that can be blocked by the expression of Bcl-2. Interestingly, inhibition of apoptosis by Bcl-2 shifts the viral infection from lytic to persistent. Other viruses, including influenza virus, Dengue virus, various herpesviruses and HIV, have also been reported to induce apoptosis as a result of cellular infections.37 Poliovirus, therefore, is not unique with regard to its ability to induce apoptosis. Poliovirus-induced apoptosis may, therefore, have an important function in the viral life cycle through limiting the host immune responses to the virus and by facilitating viral persistence.
Cleavage of Trif and Cardif appears to be a viral strategy that has been adopted by several viruses. The HCV protease NS3-4A cleaves Cardif after amino acids 508 (Figure 6), only a few amino acids before the mitochondrial membrane anchor site. In contrast, HAV protease 3ABC cleaves Cardif after amino acid Q427, next to the caspase cleavage site (D429). Although all these sites are different, their cleavage results in the complete inactivation of Cardif. The molecular rationale for this is currently not well understood. As the known Traf3 binding site (amino acids 143–147)39 is not affected by the processing of Cardif, it is more likely that successful Traf3-mediated signaling requires proximity of the mitochondria, which is impaired upon cleavage in these cases.
Model of Cardif and Trif interference by caspases and viral proteases. Both Trif and Cardif are cleaved by caspases and HCV NS3-4A. Moreover, Cardif is also cleaved by HAV 3ABC protease (it is unknown if Trif is also cleaved by HAV 3ABC). During a poliovirus infection, Cardif and MDA5 are cleaved by caspases. The caspase-targeted amino acid sequences within Cardif and Trif are indicated. CARD, caspase recruitment domain; HAV, hepatitis A virus; RHIM, RIP homotypic interaction motif; TIR, Toll-interleukin-1 receptor; TM, transmembrane domain
As a few viruses can persist in vivo and cause irreversible damage to the host, a better understanding of viral evasion strategies is required for a possible development of antiviral drugs with high selectivity. One of these viruses, HCV, efficiently cleaves both Cardif and Trif adaptors through its serine protease NS3-4A. Interestingly, NS3-4A inhibitors are currently under development, and initial trials in patients show very promising results.40 Another example of a major human health concern is poliovirus, whose infection can be devastating in humans. As MDA5 recognizes encephalomyocarditis virus picornavirus within infected cells,16, 17 it may also detect poliovirus, another picornavirus family member. MDA5 is cleaved by caspases during apoptosis,41 and during poliovirus infection, it is subjected to similar caspase-dependent, proteolytic events.34 Our data show that the MDA5 adaptor protein, Cardif, is also targeted by caspases in poliovirus-infected cells. As cleavage of Cardif resulted in the loss of antiviral activity, it is conceivable that certain viruses (such as poliovirus) evade innate immune responses through indirect (cellular protease-mediated) cleavage of crucial host molecules, thereby enhancing their replication within infected cells. On the other hand, caspase inhibition by zVAD treatment did not result in increased type-I IFN induction upon poliovirus infection, and the viral yields remained unchanged, when compared with non-treated cells.34 Hence, future studies should clarify whether caspase-mediated cleavage of Cardif and Trif is a viral strategy to counteract host responses, a host strategy to regulate innate immune responses or a host strategy to limit inflammatory gene induction during apoptosis.
Materials and Methods
Expression vectors
WT and C508A Cardif constructs, WT Trif and Luciferase reporter constructs were described.12, 31 Cardif (encoding amino acids 1–429 and 430–540) and Trif (encoding amino acids 1–281 and 290–712) deletion constructs, and Cardif (D429E) and Trif (D281E, D289E, D340E and D281, 289E (DDEE)) point mutant constructs were amplified by standard and double PCR, respectively, with Pwo Superyield polymerase (Roche), and cloned into a derivative of pCR3 (Invitrogen), in frame with a N-terminal VSV or Flag tag. The fidelity of the PCR amplifications was confirmed by sequencing.
Cell culture conditions
The human embryonic kidney (HEK) 293T, 293T-6 (a cell clone that originates from 293T, which has been selected for its high sensitivity to FasL)28 and HeLa cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated FCS. The Jurkat T-cell line, a caspase-8-deficient Jurkat clone (kindly provided by J Blenis, Harvard Medical School, Boston, MA, USA) and a caspase-9-deficient Jurkat clone, reconstituted or not with caspase-9 (both kindly provided by I Schmitz, University of Düsseldorf, Germany) were grown in RPMI (Invitrogen) supplemented with 10% heat-inactivated FCS. U-2 OS human osteosarcoma-derived cell lines that inducibly express NS3-4A upon tetracycline withdrawal have been described.42
Transfections, immunoprecipitations and immunoblots
These assays were performed as previously described.31
Reagents
Fc:FasL28 was used at 100 ng/ml unless indicated. zVAD (Alexis) was used at 25 μM. Staurosporine and etoposide (both from Alexis) were used at 250 ng/ml and 50 μM, respectively, doxorubicin (Sigma) at 20 μg/ml and camptothecin (Apotech) at 100 μM. Antibodies to caspase-8 (804-242), NS3 (1B6), Trif (AL227) Cardif (AT107), MDA5 (AT113) and RIG-I (Alme-1) were from Alexis. Anti-VSV (P5D4) was from Sigma. Anti-PARP (9542), anti-IκB (9242), anti-phospho-IκB (9246) and anti-phospho-IRF3 (4947) were from Cell Signaling. Anti-IRF3 (sc-9082) was from Santa Cruz Technology. For immunoprecipitations, antibodies to Trif (4596) were from Cell Signaling. The human RANTES ELISA kit was from R&D Systems.
Luciferase reporter assays
These assays were performed as previously described.31
Viral infections
HeLa cells were infected for the indicated time points with poliovirus (strain Mahoney) at a multiplicity of infection of 5. After the indicated time periods, the cells were harvested, lysed in RIPA buffer and analyzed by immunoblot.
For generation of cells stably expressing an shRNA-targeting Cardif, pLKO.1 vectors (Open Biosystems) were used, either empty (control) or containing the hairpin (CAAGTTGCCAACTAGCTCAAA). Cardif constructs (both WT and D429E point mutant) were rendered resistant to knockdown by a double PCR introducing three silent mutations (underlined nucleotides) in the RNAi sequence (CAAATTACCGACTAGCTCAAA). shRNAs to caspase-3 in pLKO.1 vectors were CCGAAAGGTGGCAACAGAATT (shCasp3.1) and CTAAAGGTGGTGAGGCAATAA (shCasp3.2).
Lentiviruses were produced using second-generation packaging plasmids pMD2-VSVG and pCMV-R8.91.43 293T cells were co-transfected with packaging plasmids and the shRNA-encoding plasmids. Cells were washed 18 h after transfection. Virus-containing supernatants were collected 24 h after washing, filtered and used for infection. Jurkat or 293T cells were harvested and resuspended in virus-containing supernatant. After 36 (Jurkat) or 48 h (293T), cells were washed and put under puromycin selection (Jurkat: 5 μg/ml; 293T: 2.5 μg/ml) for at least 5 days. Five different shRNAs were tested for each target protein. The expression level of the target protein was assessed by immunoblot, and the most efficiently silenced cells were selected for further experiments.
Abbreviations
- Cardif:
-
CARD adaptor-inducing interferon-β
- IFN:
-
interferon
- IPS-1:
-
interferon-β promoter stimulator 1
- IRF:
-
interferon regulatory factor
- MAVS:
-
mitochondrial antiviral signaling
- TLR:
-
Toll-like receptor
- Trif:
-
TIR domain-containing adapter inducing IFN-β
- VISA:
-
virus-induced signaling adaptor
References
Medzhitov R . Toll-like receptors and innate immunity. Nat Rev Immunol 2001; 1: 135–145.
Takeda K, Kaisho T, Akira S . Toll-like receptors. Annu Rev Immunol 2003; 21: 335–376.
O’Neill LA, Bowie AG . The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol 2007; 7: 353–364.
Alexopoulou L, Holt AC, Medzhitov R, Flavell RA . Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 2001; 413: 732–738.
Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature 2003; 424: 743–748.
Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003; 301: 640–643.
Honda K, Taniguchi T . IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol 2006; 6: 644–658.
Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA 2004; 101: 17264–17269.
Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5: 730–737.
Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 2005; 6: 981–988.
Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med 2006; 203: 1795–1803.
Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005; 437: 1167–1172.
Seth RB, Sun L, Ea CK, Chen ZJ . Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005; 122: 669–682.
Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 2006; 24: 633–642.
Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB . VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 2005; 19: 727–740.
Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA et al. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci USA 2006; 103: 8459–8464.
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441: 101–105.
Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA 2005; 102: 2992–2997.
Li XD, Sun L, Seth RB, Pineda G, Chen ZJ . Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA 2005; 102: 17717–17722.
Meylan E, Tschopp J . Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell 2006; 22: 561–569.
Yang Y, Liang Y, Qu L, Chen Z, Yi M, Li K et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci USA 2007; 104: 7253–7258.
Lin R, Lacoste J, Nakhaei P, Sun Q, Yang L, Paz S et al. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKepsilon molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J Virol 2006; 80: 6072–6083.
Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci USA 2006; 103: 6001–6006.
Takahashi K, Kawai T, Kumar H, Sato S, Yonehara S, Akira S . Roles of caspase-8 and caspase-10 in innate immune responses to double-stranded RNA. J Immunol 2006; 176: 4520–4524.
Juo P, Kuo CJ, Yuan J, Blenis J . Essential requirement for caspase-8/FLICE in the initiation of the Fas-induced apoptotic cascade. Curr Biol 1998; 8: 1001–1008.
McStay GP, Salvesen GS, Green DR . Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ 2008; 15: 322–331.
Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J . RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep 2002; 3: 1201–1208.
Holler N, Tardivel A, Kovacsovics-Bankowski M, Hertig S, Gaide O, Martinon F et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol Cell Biol 2003; 23: 1428–1440.
Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB . Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem 2004; 279: 15652–15661.
Kaiser WJ, Offermann MK . Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol 2005; 174: 4942–4952.
Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol 2004; 5: 503–507.
Ruckdeschel K, Pfaffinger G, Haase R, Sing A, Weighardt H, Hacker G et al. Signaling of apoptosis through TLRs critically involves toll/IL-1 receptor domain-containing adapter inducing IFN-beta, but not MyD88, in bacteria-infected murine macrophages. J Immunol 2004; 173: 3320–3328.
Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB . mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA 2002; 99: 637–642.
Barral PM, Morrison JM, Drahos J, Gupta P, Sarkar D, Fisher PB et al. MDA-5 is cleaved in poliovirus-infected cells. J Virol 2007; 81: 3677–3684.
Balachandran S, Thomas E, Barber GN . A FADD-dependent innate immune mechanism in mammalian cells. Nature 2004; 432: 401–405.
Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G . Viral control of mitochondrial apoptosis. PLoS Pathog 2008; 4: e1000018.
Teodoro JG, Branton PE . Regulation of apoptosis by viral gene products. J Virol 1997; 71: 1739–1746.
Best SM, Bloom ME . Caspase activation during virus infection: more than just the kiss of death? Virology 2004; 320: 191–194.
Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G et al. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 2006; 25: 3257–3263.
Lamarre D, Anderson PC, Bailey M, Beaulieu P, Bolger G, Bonneau P et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 2003; 426: 186–189.
Kovacsovics M, Martinon F, Micheau O, Bodmer JL, Hofmann K, Tschopp J . Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr Biol 2002; 12: 838–843.
Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J Virol 2000; 74: 2293–2304.
Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 1996; 272: 263–267.
Acknowledgements
We thank MC Michallet and Dan Muruve for critical readings and discussions. This study was supported by grants of the Swiss National Science Foundation and ApoTrain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by J Silke
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Rebsamen, M., Meylan, E., Curran, J. et al. The antiviral adaptor proteins Cardif and Trif are processed and inactivated by caspases. Cell Death Differ 15, 1804–1811 (2008). https://doi.org/10.1038/cdd.2008.119
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.119