Abstract
Recent evidence shows that raddeanin A (RA), an oleanane-type triterpenoid saponin extracted from Anemone raddeana Regel, exerts remarkable cytotoxicity against cancer cells in vitro and in vivo. In addition, RA has also been found to activate autophagy in human gastric cancer cells. In this study, we investigated the molecular mechanisms underlying RA-induced autophagy as well as the relationship between RA-induced autophagy and its cytotoxicity in human breast cancer cells in vitro. Treatment with RA (2–8 μmol/L) dose-dependently enhanced autophagy, as evidenced by increased LC3 levels in breast cancer cell lines T47D, MCF-7 and MDA-MB-231. Furthermore, the Akt-mTOR-eEF-2K signaling pathway was demonstrated to be involved in RA-induced activation of autophagy in the 3 breast cancer cell lines. Treatment with RA (2–10 μmol/L) dose-dependently induced apoptosis in the 3 breast cancer cell lines. Pretreatment with the autophagy inhibitor chloroquine (CQ, 20 μmol/L) significantly enhanced RA-caused cytotoxicity via promoting apoptosis. In conclusion, our results suggest that modulating autophagy can reinforce the cytotoxicity of RA against human breast cancer cells.
Similar content being viewed by others
Introduction
Raddeanin A (RA), an oleanane-type triterpenoid saponin, is extracted from the traditional Chinese herb Anemone raddeana Regel, which was used to treat rheumatism and arthritis in ancient China1. It has been reported that RA can induce apoptosis in human gastric cancer cells and colon cancer cells2,3. The anticancer activity of RA has also been demonstrated in liver cancer and cervical cancer xenograft models4.
Autophagy, also named as “type II programmed cell death”, is a highly conserved process through which cellular organelles are digested and recycled, providing energy to promote cell survival in response to metabolic stress such as nutrient deficiency and hypoxia5. Numerous studies indicate that autophagy is activated in tumor cells exposed to certain types of chemotherapy, targeted therapy and radiotherapy and that inhibiting autophagy may increase the effectiveness of concomitant treatments6. For example, targeting autophagy-mediated cell survival enhanced the anti-cancer effects of the growth factor inhibitors gefitinib and lapatinib in breast cancer cells7. Knock down of autophagy-related genes by genetic manipulation or pharmacological inhibition sensitized breast cancer cells to tamoxifen therapy8,9,10. In triple-negative breast cancer stem cells, hypoxia increased drug resistance by inducing autophagy, and molecular or chemical inhibition of autophagic pathways was able to reverse chemoresistance11. Thus, modulation of autophagy is a promising new approach to cancer treatment.
Eukaryotic elongation factor 2 kinase (eEF-2K), a calcium/calmodulin-dependent kinase, phosphorylates its only known substrate, eEF2, on Thr56, and terminates peptide elongation by impairing the ability of eEF2 to transfer peptidyl-tRNA from the ribosomal A to P site12,13. eEF-2K is overexpressed and constitutively activated in several types of cancer, including breast cancer, and contributes to cell proliferation14. The activity of eEF-2K is negatively modulated by mTOR, a key regulator of autophagy, which blocks autophagy by inhibiting the association between Atg1 and Atg13 or by inactivating ULK1 through phosphorylation15,16,17. We have reported that eEF-2K can act as a positive regulator of autophagy under metabolic or therapeutic stresses, including nutrient deprivation, growth factor inhibition, ER stress and Akt inhibition7,18.
More recently, Teng et al found that RA activates autophagy in human gastric cancer cells19. However, the exact molecular mechanisms regulating RA induction of autophagy are still unclear. In this study, we demonstrated that RA induced autophagy in human breast cancer cells and that the Akt-mTOR-eEF-2K signaling pathway is involved in RA activation of autophagy. Inhibition of autophagy enhanced the cytotoxicity of RA against breast cancer cells by promoting apoptosis.
Materials and methods
Reagents and antibodies
Raddeanin A was a generous gift from China Pharmaceutical University. Chloroquine (CQ) was purchased from Sigma-Aldrich. MK2206 was a gift from APE×BIO.
Rabbit monoclonal antibodies against LC3, phospho-p70S6K (T389), p70S6K, phospho-Akt (S473), Akt, phospho-eEF2 (T56), eEF2, eEF-2K, cleaved caspase-3, PARP, Bcl-xL, Mcl-1 and Bcl-2 were purchased from Cell Signaling Technologies. Anti-β-Actin was purchased from Proteintech.
Cell lines and cell culture
MCF-7 and T47D cells were cultured in DMEM high glucose medium supplemented with 10% FBS (Gibco), and maintained at 37 ˚C with 5% CO2 in a humidified atmosphere. MDA-MB-231 cells were grown in L-15 medium supplemented with 10% FBS, and cultured at 37 ˚C with 100% air.
Western blot analysis
Cells were seeded in 6-well-plates. After treatment, cells were lysed by RIPA buffer (Beyotime, Haimen, China) supplemented with a protease inhibitor cocktail and a phosphatase inhibitor cocktail (Selleck). Protein concentrations were quantified with a BCA protein assay kit (Beyotime, Haimen, China). The proteins were separated by 10%–12% SDS-PAGE and transferred to a PVDF membrane. The PVDF membrane was incubated with primary antibody in 5% BSA/PBS buffered with Tween 2.0 at 4 ˚C overnight. Membranes were then incubated with anti-rabbit or anti-mouse secondary antibodies at room temperature for 1 h. Detection was accomplished by chemiluminescence using an ECL reagent.
Cell viability assay
Cells were seeded at 7×103 cells per well in 96-well-plates, and treated with a series of RA concentrations for 48 h. At the end of the treatment, 10 μL of CCK8 reagent (Biotool) was added to each well, and the cells were incubated for 2 h. At the end of this incubation, the absorbance at 450 nm wavelength was measured.
siRNA and plasmid transfection
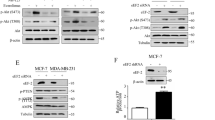
siRNA targeting eEF-2K was purchased from Ribobio. Non-targeting siRNA was used as a control. Transfection of siRNA was accomplished according to the manufacturer's protocol. In brief, cells in the exponential phase of growth were plated in 6-well tissue culture plates at 1×105 cells per well, grown for 24 h, then transfected with siRNA using Lipofectamine 2000 (Invitrogen) and OPTI-MEM reduced serum medium. For plasmid transfection, cells were transfected with a mixture of GFP-LC3 plasmid and Lipofectamine 2000 (Invitrogen) in OPTI-MEM reduced serum medium.
Hoechst 33258 staining
After treatment, the cells were washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature, and then incubated with Hoechst 33258 (Beyotime) for 30 min at 37 ˚C. Apoptotic cells were identified by the presence of nuclear shrinkage and chromatin condensation and fragmentation.
Caspase-3 activity assay
The activity of caspase-3 was evaluated using caspase-3 activity kit (Beyotime). Briefly, 50 μL cell lysates were incubated with 50 μL reaction buffer containing 0.2 mmol/L caspase-3 substrate (Ac-DEVD-pNA) in 96-well tissue plates for 2 h at 37 ˚C. At the end of the incubation, the absorbance at 405 nm wavelength was measured.
Statistical analysis
A 2-sample t-test was used to analyze differences between samples treated with RA alone and samples treated with RA in combination with an autophagy inhibitor. Differences between groups were considered statistically significant at P<0.05.
Results
RA induces autophagy in breast cancer cells
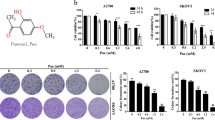
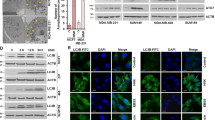
The breast cancer cell lines T47D, MCF-7 and MDA-MB-231 were treated with RA, and the level of LC3, a hallmark of autophagy, was measured. As shown in Figure 1A, RA increased the expression of LC3-II in a dose- and time-dependent manner. We further monitored autophagic flux and found that LC3-II accumulated in the presence of the autophagy inhibitor chloroguine (CQ), indicating that autophagic flux was enhanced by RA (Figure 1B). To further confirm the stimulatory effect of RA on autophagy, we transfected MCF-7 cells with a GFP-LC3 plasmid and observed an increase in GFP-LC3 puncta in the cells treated with the compound (Figure 1C). The above results demonstrate that RA can induce autophagy in breast cancer cells.
RA induces autophagy in breast cancer cells. (A) T47D, MCF-7 or MDA-MB-231 cells were treated with a series of concentrations of RA for 24 h or with 8 μmol/L RA for different durations. The levels of LC3 were measured by Western blot. β-Actin was used as a loading control. (B) T47D or MCF-7 cells were pretreated with 20 μmol/L CQ for 1 h, followed by treatment with RA for 24 h. The level of LC3 was measured by Western blot. β-Actin was used as a loading control. (C) MCF-7 cells were transfected with a GFP-LC3 plasmid, followed by treatment with 5 μmol/L RA for 12 h. GFP-LC3 puncta were observed at 40×magnification under an inverted fluorescence microscope.
The Akt-mTOR-eEF-2K pathway is involved in the activation of autophagy induced by RA
Next, we explored the pathways that mediate the activation of autophagy induced by RA. We have identified eEF-2K as a regulator of autophagy under stress; therefore, we sought to determine whether this kinase could also regulate autophagy induced by RA. As shown in Figure 2A, the phosphorylation of eEF2, the only known substrate of eEF-2K, was increased in a concentration- and time-dependent manner in the cells treated with RA, indicating that eEF-2K is activated by this compound. To demonstrate the role of eEF-2K in the activation of autophagy by RA, we transfected cells with eEF-2K siRNA and measured autophagic activity. Figure 2B shows that when eEF-2K was silenced, there was a decrease in the amount of LC3-II compared with that in the cells in which the kinase was not silenced, indicating that the autophagic response to RA treatment was blunted by inactivation of eEF-2K. To explore the pathway that mediates the activation of eEF-2K by RA, we examined mTOR activity, which acts as a negative regulator of eEF-2K by phosphorylation. Figure 2C shows that RA decreased mTOR activity in a dose- and time-dependent manner, as evidenced by a decrease in the level of p-S6K, the downstream effector of mTOR. We next measured the activity of Akt, a positive upstream regulator of mTOR, and found that the level of p-Akt was downregulated in cells treated with this compound (Figure 2C). To further validate the relationship between eEF-2K and Akt, we treated T47D or MCF-7 cells with MK2206, a small molecule allosteric inhibitor of Akt, and found that MK2206 increased the level of p-eEF2 in a concentration-dependent manner (Figure 2D), indicating that inhibition of Akt activated eEF-2K. These results suggest that induction of autophagy by RA is mediated via the Akt-mTOR-eEF-2K pathway.
The Akt-mTOR-eEF-2K pathway is involved in the activation of autophagy induced by RA. (A) T47D, MCF-7 or MDA-MB-231 cells were treated with a series of concentrations of RA for 24 h or with 8 μmol/L RA for different durations. After treatment, the levels of phospho-eEF2 (T56) and eEF2 were measured by Western blot. β-Actin was used as a loading control. (B) MDA-MB-231 or T47D cells were transfected with a non-targeting siRNA or an siRNA targeting eEF-2K, followed by treatment with 6 μmol/L RA for 24 h. The levels of eEF-2K, phospho-eEF2 (T56), eEF2, and LC3 were measured by Western blot. β-Actin was used as a loading control. (C) T47D, MCF-7 or MDA-MB-231 cells were treated with a series of concentrations of RA for 24 h or with 8 μmol/L RA for different durations. After treatment, the levels of phospho-p70S6K (T389), p70S6K, phospho-Akt (S473) and Akt were measured by Western blot. β-Actin was used as a loading control. (D) T47D or MCF-7 cells were treated with a series of concentrations of MK2206 for 24 h. After treatment, the levels of phospho-eEF2 (T56), eEF2, phospho-Akt (S473) and Akt were measured by Western blot. β-Actin was used as a loading control.
RA induces apoptosis in breast cancer cells
It has been reported that RA can inhibit cell proliferation and induce apoptosis in multiple cancer cell lines, including gastric cancer and colon cancer cell lines1,3. Accordingly, we examined the effect of RA on apoptosis in breast cancer cells. Figure 3A shows that treatment of breast cancer cells with RA resulted in a dose- and time- dependent downregulation of the levels of anti-apoptotic Bcl-2 family proteins, including Bcl-xL, Mcl-1, and Bcl-2. Marked apoptotic morphologic alterations, including cell shrinkage, membrane blebbing and apoptosis bodies, were observed in breast cancer cells treated with RA (Figure 3B). The activation of caspase-3 and PARP is involved in Bcl-2 family-mediated induction of apoptosis. Figure 3C shows that in T47D and MCF-7 cells, the levels of cleaved caspase-3 and PARP increased in a concentration-dependent manner after treatment with RA. Furthermore, Hoechst 33258 staining revealed typical nuclear morphological changes of apoptosis in RA-treated cancer cells, characterized by nuclear shrinkage and chromatin condensation and fragmentation (Figure 3D).
RA induces apoptosis in breast cancer cells. (A) T47D, MCF-7, or MDA-MB-231 cells were treated with a series of concentrations of RA for 24 h or with 8 μmol/L RA for different durations. After treatment, the levels of Bcl-xL, Mcl-1 and Bcl-2 were measured by Western blot. β-Actin was used as a loading control. (B) T47D cells were treated with various concentrations of RA for 24 h, and cellular morphological changes were observed at 10×magnification under a microscope. Arrows indicate apoptosis bodies. The numbers of apoptotic cells were counted in more than 3 fields, with at least 100 cells for each group. The results are displayed as the mean±SD of 3 experiments; **P<0.01 vs the control group. (C) T47D or MCF-7 cells were treated with a series of concentrations of RA for 24 h. After treatment, the levels of cleaved caspase-3 and PARP were measured by Western blot. β-Actin was used as a loading control. (D) MCF-7 cells were treated with 10 μmol/L RA for 24 h. Apoptotic morphology of nuclei were evaluated with Hoechst 33258 staining analysis and observed at 20×magnification under an inverted fluorescence microscope. Arrows indicate nuclear shrinkage, chromatin condensation and fragmentation. Scale bar=50 μm.
Suppression of autophagy enhances the cytocidal effect of RA by promoting apoptosis in breast cancer cells
To further understand the relationship between autophagy activation and apoptosis in RA-treated cells, we measured the effect of autophagy on apoptosis. Figure 4A shows that inhibition of autophagy by CQ, a chemical inhibitor of autophagy, augmented apoptosis compared to treatment with RA alone, as determined by the downregulation of Bcl-2 and Mcl-1, and increased expression of cleaved caspase-3 and PARP. Inhibition of autophagy increased RA-induced apoptotic activity, and this was also confirmed by the observed increase in caspase-3 activity (Figure 4B). We further determined the effects of autophagy on breast cancer cell viability upon treatment with RA and observed that the cytocidal activity of RA was also markedly increased in cells simultaneously treated with CQ (Figure 4C), suggesting that inhibition of autophagy can sensitize tumor cells to this compound.
Suppression of autophagy enhances the cytotoxic effect of RA by promoting apoptosis in breast cancer cells. (A) T47D or MCF-7 cells were pretreated with 20 μmol/L CQ for 1 h, followed by treatment with RA for 24 h. After treatment, the levels of Bcl-2, Mcl-1, LC3, cleaved caspase-3 and PARP were measured by Western blot. β-Actin was used as a loading control. (B) MDA-MB-231 cells were pretreated with 20 μmol/L CQ for 1 h, followed by treatment with 4 μmol/L RA for 12 h. After treatment, caspase-3 activity was measured with a kit and normalized to the control. The results are displayed as the mean±SD of 3 experiments; *P<0.05, **P<0.01, RA vs the control group, RA vs RA+CQ. (C) MCF-7, T47D or MDA-MB-231 cells were treated with a series of concentrations of RA for 48 h in the absence or presence of 20 μmol/L CQ. After treatment, cell viability was measured using a CCK8 reagent. The results are displayed as the mean±SD of triplicate measurements from one of three identical experiments; *P<0.05, **P<0.01, t-test, RA vs RA+CQ.
Discussion
Because of the successful discovery of anticancer drugs from natural products, such as vinca alkaloids and paclitaxel, medicinal plants are considered to be an important source of drugs for the treatment of different cancer types. As a natural bioactive substance, RA has been demonstrated to exhibit anticancer activity by inducing apoptosis in cancer cells, including gastric cancer, colon cancer and hepatocellular carcinoma1,3. A better understanding of the mechanisms of tumor cell death induced by RA could facilitate the development of this compound as an anticancer agent. In this study, we found that RA can induce autophagy and apoptosis in breast cancer cells. In addition, this study reveals the signaling pathway underlying the activation of autophagy and the relationship between autophagy and apoptosis in cancer cells exposed to RA.
Numerous studies indicate that autophagy is activated in tumor cells exposed to certain types of chemotherapy (eg, doxorubicin, temozolomide, etoposide, etc) and targeted therapy, and protects the viability of tumor cells by providing nutrients and energy20. Autophagy inhibitors, such as chloroquine and hydroxychloroquine, can augment the anticancer activity of cyclophosphamide in Myc-driven lymphoma21. Inhibiting autophagy also sensitized gastrointestinal stromal tumor cells to imatinib treatment, both in vitro and in vivo22. Therefore, suppression of autophagy may exert therapeutic effects by promoting cell death. Extensive investigation has focused on therapeutic targeting of the autophagy pathway as a new anticancer strategy. Here, we found that the activation of autophagy induced by RA was cytoprotective in tumor cells, since suppression of autophagy promoted apoptotic cell death and enhanced the cytotoxicity of the compound. These results indicate that targeting autophagy may be exploited to enhance the anticancer effects of RA.
We have previously reported that eEF-2K can act as a positive regulator of autophagy in cancer cells in response to various stresses. Here, we demonstrated that RA can activate eEF-2K and that knockdown of this kinase blocked the activity of autophagy induced by RA. This study further underscores the importance of this kinase as a regulator of autophagy. To explore the pathways underlying RA-mediated activation of eEF-2K, we tested the role of mTOR, which is a key regulator of cell growth and proliferation and has been demonstrated to be a negative regulator of autophagy. eEF-2K can be inhibited via phosphorylation at Ser78 and Ser366 by mTOR16,23. We found that RA downregulated the activity of mTOR, as determined by the decreased level of p-S6K. We further found that RA inhibited the activity of Akt, an upstream regulator of mTOR. Therefore, the activation of eEF-2K by RA is mediated through the Akt-mTOR pathway.
Based on these findings, we report here that the activation of autophagy in breast cancer cells by RA is mediated by the Akt-mTOR-eEF-2K signaling pathway and that inhibition of autophagy promotes apoptosis and sensitizes tumor cells to the cytotoxic effects of the compound.
Author contribution
Yan CHENG, Xiao-jun XU and Song-shu XIAO designed, conceived the experiments; Yi-di GUAN, Shi-long JIANG, Pian YU and Mei WEN performed the experiments; Yi ZHANG provided technical and material support; Yi-di GUAN analyzed the data, wrote the manuscript; Yan CHENG reviewed and edited the manuscript.
References
Li JN, Yu Y, Zhang YF, Li ZM, Cai GZ, Gong JY . Synergy of Raddeanin A and cisplatin induced therapeutic effect enhancement in human hepatocellular carcinoma. Biochem Biophys Res Commun 2017; 485: 335–41.
Gu G, Qi H, Jiang T, Ma B, Fang Z, Xu H, et al. Investigation of the cytotoxicity, apoptosis and pharmacokinetics of Raddeanin A. Oncol Lett 2017; 13: 1365–9.
Xue G, Zou X, Zhou JY, Sun W, Wu J, Xu JL, et al. Raddeanin A induces human gastric cancer cells apoptosis and inhibits their invasion in vitro. Biochem Biophys Res Commun 2013; 439: 196–202.
Wang MK, Ding LS, Wu FE . Antitumor effects of raddeanin A on S180, H22 and U14 cell xenografts in mice. Ai Zheng 2008; 27: 910–3.
Cheng Y, Ren X, Hait WN, Yang JM . Therapeutic targeting of autophagy in disease: biology and pharmacology. Pharmacol Rev 2013; 65: 1162–97.
Piya S, Andreeff M, Borthakur G . Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy 2017; 13: 214–5.
Cheng Y, Li H, Ren X, Niu T, Hait WN, Yang J . Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One 2010; 5: e9715.
Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat 2008; 112: 389–403.
Thomas S, Thurn KT, Bicaku E, Marchion DC, Munster PN . Addition of a histone deacetylase inhibitor redirects tamoxifen-treated breast cancer cells into apoptosis, which is opposed by the induction of autophagy. Breast Cancer Res Treat 2011; 130: 437–47.
Cook KL, Warri A, Soto-Pantoja DR, Clarke PA, Cruz MI, Zwart A, et al. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clin Cancer Res 2014; 20: 3222–32.
Bousquet G, El Bouchtaoui M, Sophie T, Leboeuf C, de Bazelaire C, Ratajczak P, et al. Targeting autophagic cancer stem-cells to reverse chemoresistance in human triple negative breast cancer. Oncotarget 2017; 8: 35205–21.
Cheng Y, Yan L, Ren X, Yang JM . eEF-2 kinase, another meddler in the “yin and yang” of Akt-mediated cell fate? Autophagy 2011; 7: 660–1.
Kenney JW, Moore CE, Wang X, Proud CG . Eukaryotic elongation factor 2 kinase, an unusual enzyme with multiple roles. Adv Biol Regul 2014; 55: 15–27.
Parmer TG, Ward MD, Yurkow EJ, Vyas VH, Kearney TJ, Hait WN . Activity and regulation by growth factors of calmodulin-dependent protein kinase III (elongation factor 2-kinase) in human breast cancer. Br J Cancer 1999; 79: 59–64.
Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30: 1049–58.
Wang X, Regufe da Mota S, Liu R, Moore CE, Xie J, Lanucara F, et al. Eukaryotic elongation factor 2 kinase activity is controlled by multiple inputs from oncogenic signaling. Mol Cell Biol 2014; 34: 4088–103.
Fingar DC, Salama S, Tsou C, Harlow E, Blenis J . Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 2002; 16: 1472–87.
Wu H, Yang JM, Jin S, Zhang H, Hait WN . Elongation factor-2 kinase regulates autophagy in human glioblastoma cells. Cancer Res 2006; 66: 3015–23.
Teng YH, Li JP, Liu SL, Zou X, Fang LH, Zhou JY, et al. Autophagy protects from Raddeanin A-induced apoptosis in SGC-7901 human gastric cancer cells. Evid Based Complement Alternat Med 2016; 2016: 9406758.
Lei Y, Zhang D, Yu J, Dong H, Zhang J, Yang S . Targeting autophagy in cancer stem cells as an anticancer therapy. Cancer Lett 2017; 393: 33–9.
Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Investig 2007; 117: 326–36.
Janku F, McConkey DJ, Hong DS, Kurzrock R . Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011; 8: 528–39.
Fu LL, Xie T, Zhang SY, Liu B . Eukaryotic elongation factor-2 kinase (eEF2K): a potential therapeutic target in cancer. Apoptosis 2014; 19: 1527–31.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No 81422051, 81472593 and 31401208).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Guan, Yd., Jiang, Sl., Yu, P. et al. Suppression of eEF-2K-mediated autophagy enhances the cytotoxicity of raddeanin A against human breast cancer cells in vitro. Acta Pharmacol Sin 39, 642–648 (2018). https://doi.org/10.1038/aps.2017.139
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2017.139
Keywords
This article is cited by
-
Saponins as cytotoxic agents: an update (2010–2021). Part II—Triterpene saponins
Phytochemistry Reviews (2023)
-
Raddeanin A promotes autophagy-induced apoptosis by inactivating PI3K/AKT/mTOR pathway in lung adenocarcinoma cells
Naunyn-Schmiedeberg's Archives of Pharmacology (2023)
-
Eukaryotic elongation factor-2 kinase regulates the cross-talk between autophagy and pyroptosis in doxorubicin-treated human melanoma cells in vitro
Acta Pharmacologica Sinica (2019)
-
Tubeimoside-1, a triterpenoid saponin, induces cytoprotective autophagy in human breast cancer cells in vitro via Akt-mediated pathway
Acta Pharmacologica Sinica (2019)