Abstract
Both iptakalim (Ipt) and natakalim (Nat) activate the SUR2B/Kir6.1 channel, an ATP-sensitive potassium channel (KATP) subtype, with high selectivity. In this study we investigated the therapeutic effects of Ipt and Nat against isoproterenol-induced chronic heart failure (ISO-CHF) in rats, and demonstrated a new therapeutic approach to the treatment of CHF through activation of the SUR2B/Kir6.1 channel in endothelial cells. In ISO-CHF rats, oral administration of Nat (1, 3, 9 mg·kg−1·d−1) or Ipt (3 mg·kg−1·d−1) for 60 days significantly improved cardiac dysfunction, reversed cardiac remodeling, significantly attenuated the pathological increases in BNP levels, and improved endothelial dysfunction by adjusting the balance between endothelin and NO systems. The therapeutic effects of Nat were prevented by the selective KATP blocker glibenclamine (Gli, 50 mg·kg−1·d−1), confirming that these effects were mediated through activation of the SUR2B/Kir6.1 channel in endothelial cells. The molecular mechanisms underlying the therapeutic effects of Nat were further addressed using proteomic methods. We identified 724 proteins in the plasma of ISO-CHF rats; 55 proteins were related to Nat. These differentially expressed proteins were mainly involved in single-organism processes and the regulation of biological quality relative to CHF, including proteasome (Psm) and ATP protein clusters. We screened out PRKAR2β, GAS6/eNOS/NO and NO/PKG/VASP pathways involved in the amelioration of CHF among the 24 enriched pathways. We further confirmed 6 protein candidates, including PRKAR2β, GAS6 and VASP, which were involved in the endothelial mechanisms, and ATP, TIMP3 and AGT, which contributed to its cardiovascular actions. This study demonstrates a new pharmacological approach to the treatment of CHF through activation of the SUR2B/Kir6.1 channel in endothelial cells, and that the eNOS/VASP pathways are involved in its signaling mechanisms.
Similar content being viewed by others
Introduction
Chronic heart failure (CHF) is the leading lethal disease among cardiovascular diseases worldwide. Hypertensive heart diseases, myocardial infarction (MI), arrhythmia, and other common cardiovascular diseases are eventually exacerbated into CHF due to pressure overload (PO), myocardial damage, or weakened myocardial contraction1,2,3. Endothelial dysfunction plays an important role in the development of CHF induced by PO (PO-CHF), MI (MI-CHF), and tachycardia, which are mimicked by the administration of isoproterenol (ISO, ISO-CHF), as well as hypertensive cardiovascular remodeling (HCVR)4,5,6. Serial experiments from our research centers have suggested that both iptakalim (Ipt) and its new derivative natakalim (Nat), new ATP-sensitive potassium channel (KATP) openers (KCOs), could activate the SUR2B/Kir6.1 channel with high selectivity. We investigated the effects of both Ipt and Nat in PO-CHF and explored the effects of Nat in MI-CHF and ISO-CHF. Our results in the independent experiments indicated that both Ipt and Nat could restore endothelial function and improve PO-CHF, MI-CHF, ISO-CHF, and HCVR7,8,9,10.
To further illustrate this new anti-CHF therapeutic pathway, which is mediated by SUR2B/Kir6.1 channel opening, we attempted to answer the following questions: (1) whether the highly selective SUR2B/Kir6.1 channel openers Nat and Ipt have equivalent effectiveness against CHF under the same experimental conditions, (2) whether the highly selective KATP blocker (KCB) glibenclamide (Gli) blocks the cardiovascular effects of Nat in vivo, (3) whether the SUR2B/Kir6.1 channel openers and the angiotensin converting enzyme inhibitor (ACEI), lisinopril (Lis), have different mechanisms of action against ISO-CHF, and (4) what molecules are involved in the molecular mechanisms underlying the SUR2B/Kir6.1 channel pathway actions against ISO-CHF. In this study, we compared the therapeutic effects of Nat and Ipt in ISO-CHF and found that their effects were primarily prevented by the KCB Gli in vivo. To achieve better understanding of how Nat affects the molecular processes acting against CHF, isobaric tags for relative and absolute quantification (iTRAQ), proteomics analysis, enzyme-linked immunosorbent assay (ELISA), and Western blot analyses were performed to generate novel insights into the involved molecular mechanisms.
The objective of these investigations was to provide experimental evidence for the new therapeutic pathway against CHF mediated by the SUR2B/Kir6.1 channel in endothelial cells. Furthermore, we sought to obtain a better understanding of the detailed mechanisms against CHF.
Materials and methods
Drugs and reagents
Ipt and Nat were synthesized by Nhwa Thad Pharmaceutical Co, Ltd, (Xuzhou, China), and their chemical structures were clear. ISO, Gli, and Lis were purchased from Sigma-Aldrich (St Louis, MO, USA). Vasodilator-stimulated phosphoprotein (VASP) monoclonal antibody was purchased from Abcam (Cambridge, MA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was purchased from BioEasy (Bioeasytech, Beijing, China). The EA.hy926 human umbilical vein endothelial cell (HUVEC) line used in the present study was obtained from the American Type Culture Collection (ATCC). All other chemicals and materials were obtained from local commercial sources.
Animal model and protocol
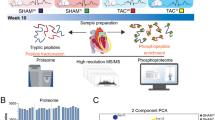
All experiments were conducted with adherence to the NIH Guide for the Care and Use of Laboratory Animals and were approved by the local animal care and use committee. The experiments were performed on 150 healthy adult male, SPF Wistar rats (weight, 200±20 g). The animals were divided into a control group (n=14) and 8 other groups (n=17/group): model (ISO), 3 Nat treatment groups (1, 3, and 9 mg·kg−1·d−1), Gli treatment group, Gli combined with Nat (3 mg·kg−1·d−1) treatment group, Ipt 3 mg·kg−1·d−1 treatment group, and ACEI Lis 15 mg·kg−1·d−1 treatment group. In the 8 test groups, rats were administered ISO (85 mg·kg−1·d−1, subcutaneous, sc) for 7 consecutive days, while the control group rats received an equal volume of physiological saline. The rats were further assigned at random to receive either Nat (1, 3, or 9 mg·kg−1·d−1, orally, po), Ipt (3 mg·kg−1·d−1, po), Lis (15 mg·kg−1·d−1, po), or an equal volume of physiological saline from the day of the ISO injection up to d 60. The Gli combined with Nat 3 mg·kg−1·d−1 treatment group rats were administered Gli (50 mg·kg−1·d−1, po) for 1 h before they were administered Nat. Nat, Ipt, Lis, and vehicle (<2 mL/kg) were orally administered once a day.
Hemodynamics index
On d 60, all the animals were weighed and anaesthetized with pentobarbital (45 mg/kg, intraperitoneally, ip); the right carotid and femoral arteries were cannulated with a polyethylene catheter connected to a Statham transducer. Then, the catheter was inserted along the right coronary artery into the left ventricle, and the signals were recorded on an eight-channel direct-writing oscillograph (RM-6000, Nihon Kohden Kogyo Co, Ltd, Japan) and digitally sampled (1 kHz) on a computer equipped with an analog to digital interface (SMUP-PC Bioanalysis System, Japan). The parameters of heart rate, cardiac systolic and diastolic function, systolic and diastolic blood pressure, and mean arterial blood pressure were monitored and recorded at 0, 5, 10, and 20 min, respectively.
Histological analysis, transmission electron microscopy (TEM), and immunohistological staining
Thereafter, the thoracic cavity was opened to expose the beating heart. Hearts were rapidly removed, rinsed in ice-cold 0.9% sodium chloride (NaCl) solution, dried (fluids were removed), weighed, and then photographed. The hearts were subsequently fixed by immersion in neutral 10% buffered formalin, and then paraffin sections (5 mm) were cut in accordance with the previously mentioned methods8. The myocardial samples were routinely fixed in 2.5% glutaraldehyde in 0.1 mol/L phosphate buffer (pH 7.3) and post-fixed in buffered 1% osmium tetroxide. TEM (Hitachi H-7650, Japan) and immunohistological staining were performed in accordance with previously reported methods8,9.
N-terminal prohormone brain natriuretic peptide (NT-proBNP), nitric oxide (NO), and endothelin-1 (ET-1) measurement
After collection and centrifugation, the blood samples were stored in liquid nitrogen until assay. Because of its instability in physiological solutions, most NO was rapidly converted to NO2− and further to NO3−. Therefore, the levels of NO2−/NO3− in the serum were measured with an NO detection kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer′s instructions. The plasma N-terminal prohormone brain natriuretic peptide (NT-proBNP) and endothelin-1 (ET-1) in the plasma were assayed using a commercial rat ELISA assay kit (Pulilai Biotechnology Company, Beijing, China), according to the instructions provided by the manufacturer.
Total RNA extraction and quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from the cardiac tissue by RNAzol RT RNA isolation reagent (E01010A, GeneCopoeia), and the first-strand cDNA was produced using RevertAidTM first-strand cDNA synthesis kit (TOYOBO, Japan) according to the manufacturer′s instructions. The amplification was performed on a Bio-Rad iQ5 system, using the primers listed in Table 1. All primers were synthesized by Invitrogen (The Netherlands). Levels of mRNA were subsequently normalized to GAPDH mRNA levels, and the relative mRNA expression levels of the genes were calculated using the 2−ΔΔCT method. The mean values were calculated from more than triplicate qRT-PCR reactions.
Protein preparation, digestion, and iTRAQ labeling
Individual plasma samples were collected from each group according to the experimental design (Figure 1). Eight of the highly abundant proteins in the plasma samples were depleted using Proteo-MinerTM kits (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer′s protocol. An aliquot of the supernatant was collected for protein concentration determination using the Bradford method. The total protein (100 μg) was accurately extracted from each sample solution and then digested with trypsin gold (Promega, Madison, WI, USA). After trypsin digestion, the samples were labeled with the iTRAQ tags as follows (Figure 1): two independent biological triplicates (model group labeled with reagents 117, 119, and 121; Nat 3 mg·kg−1·d−1 group labeled with reagents 113, 115, and 116), and one biological duplicate (control group labeled with reagents 114 and 118). The peptides were labeled with the isobaric tags, following which the labeled peptide mixtures were pooled and dried by vacuum centrifugation.
Experimental design of iTRAQ-based quantitative proteomics analysis. The model group was administered ISO, and Nat was given at a dose of 3 mg·kg−1·d−1.
Strong cation exchange (SCX) fractionation and liquid chromatography-electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) analysis based on triple time-of-flight (TOF) 5600
The labeled peptide mixtures were separated by SCX chromatography with an LC-20AB high-performance liquid chromatography (HPLC) pump system (Shimadzu, Kyoto, Japan). The mixtures were loaded onto a 4.6 mm×250 mm Ultremex SCX column containing 5-μm particles (Phenomenex) and eluted at a predetermined flow rate with a gradient of buffer A [25 mmol/L NaH2PO4 in 25% acetonitrile (ACN), pH 2.7] and buffer B (25 mmol/L NaH2PO4, 1 mol/L KCl in 25% ACN, pH 2.7). The elution was monitored by measuring the absorbance at 214 nm. The eluted peptides were pooled into 20 fractions, desalted with a Strata X C18 column (Phenomenex), and vacuum-dried11,12. The SCX fraction was analyzed by an LC-20AD nano HPLC (Shimadzu, Kyoto, Japan). The data were acquired using a Triple TOF 5600 System (AB SCIEX, Concord). The mass spectrometer (MS) was operated in an information-dependent acquisition mode. Peptides were selected for tandem mass spectrometry (MS/MS) analysis according to the accumulation times11,12.
Data processing and analysis
The raw data files acquired from the Orbitrap were converted into Mascot generic format (MGF) files using the Proteome Discoverer 1.2 (PD1.2, Thermo). The MGF files were searched, and the proteins were identified by using Mascot search engine (Matrix Science, UK, version 2.3.02). The identification of each confident protein required at least one unique peptide. For protein quantification, each protein was required to contain at least two unique spectra. The quantitative protein ratios were weighed and normalized to the median ratio in Mascot13. We only used ratios with P<0.05, and only fold changes≥1.2 or ≤0.8 were considered significant.
For bioinformatics analysis, the functional annotations of the identified proteins were conducted using the blast-GO program against the non-redundant protein database (NR; NCBI). The Clusters of Orthologous Groups of proteins (COG) and the Kyoto Encyclopedia of Genes and Genomes (KEGG, (http://www.genome.jp/kegg/) databases were used to classify and group the identified proteins. The protein interactions were explored using web-based bioinformatics tools, a search tool for the retrieval of interacting genes/proteins (STRING v 9.0).
Verification assays by ELISA
The levels of metalloproteinase inhibitor 3 (TIMP3), angiotensinogen (AGT), cyclic adenosine monophosphate (cAMP)-dependent protein kinase (PKA) type II-beta regulatory subunit (PRKAR2β), growth arrest specific gene 6 (GAS6), and VASP were measured in plasma samples from the control, model, and Nat 3 mg groups (9 subjects from each group). The measurements were carried out using a rat′s ELISA quantification kit (USCN Life Sciences, Wuhan, China) according to the manufacturer′s instructions. The plasma levels of TIMP3, AGT, PRKAR2β, GAS6, and VASP were represented using a scatter plot.
Cell culture, drug treatment, and NO measurement
The EA.hy926 cell line was cultured in DMEM/high glucose (Thermo, USA) supplemented with 10% fetal bovine serum (Gibco). The cells were maintained at 37 °C, exposed to an atmosphere of 5% CO2, seeded in 6-well plates and subsequently prepared for the drug administration when they had grown to 80% confluence.
Nat was dissolved in serum-free medium, and then the cells were treated with 10−5 mol/L Nat for 72 h before treatment with 10−3 mol/L L-NAME for 2 h. The control group was treated without Nat and L-NAME. To assess the effects of Nat and L-NAME on NO production, the NO in the media of the different groups was measured using an NO detection kit (Griess reagent) according to the manufacturer′s instructions.
Western blot
The cell lysates were prepared in ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer, and the protein content was determined by bicinchoninic acid (BCA) protein assay kit. Fifty micrograms of each protein sample was separated using 10% SDS-PAGE gels and subsequently transferred to polyvinylidene fluoride (PVDF) membranes. The VASP and GAPDH (internal control) were detected by incubating the membranes with rabbit anti-VASP monoclonal and anti-GAPDH antibodies (1:1000 and 1:3000), respectively. The PVDF membranes were subsequently probed with the secondary antibody (1:3000). Then, the antigen-antibody complexes were detected using an enhanced chemiluminescent reagent, and the Image-Pro Plus software was used for the densitometric analysis.
Statistical analysis
The statistical analyses were conducted using SPSS 17.0 software (Chicago, IL, USA). The data are expressed as the mean±standard deviation (SD). The two-tailed Student′s t-test was used to compare 2 independent groups containing normally distributed data, and one- or two-way ANOVA followed by the Student-Newman-Keuls test (SNK) was used to account for multiple comparisons. P values of less than 0.05 were considered statistically significant.
Results
ISO-CHF improvement
Body weight, blood pressure, and heart rate
The basic data are shown in Table 2. The body weights, blood pressures, and heart rates of the ISO rats decreased compared to those of the control rats. Furthermore, treatment with Nat and Ipt restored these basic indices, while the KCB Gli did not affect the actions of ISO. However, the effects of Nat could be blocked by treatment with Gli in vivo.
Hemodynamics
The hemodynamic data obtained just before euthanasia are shown in Table 3. The systolic cardiac parameters, including LVSP, +dp/dtmax, Vpm, and Vmax, and the diastolic cardiac parameter –dp/dtmax, were significantly decreased in ISO-CHF rats (P<0.05), while the diastolic cardiac parameter LVEDP was significantly increased (P<0.05). Nat could restore cardiac parameters to control levels in a dose-dependent manner. At the same dosage (3 mg), the effects of Nat and Ipt were equivalent. Nat effects could be antagonized by Gli in vivo.
Cardiac remodeling
As shown in Figure 2A, the pathological examination of the myocardial tissue in control group showed neat cardiac muscle fibers, while edema, degeneration, necrosis, and cardiac hemorrhage were not observed. In contrast, broken left ventricular myocardial fibers (MF), necrosis, and inflammatory cell infiltration could be observed in the ISO group. The results showed that Nat could improve these pathological changes dose-dependently and that the therapeutic effects of Nat and Ipt were equivalent at the same dosage of 3 mg·kg−1·d−1. Both drugs could restore the normal cardiac conditions, and Nat effects could be prevented by Gli in vivo.
Effects of natakalim on cardiac structure in the ISO-CHF rats. (A) Myocardial tissue stained with hematoxylin and eosin (H&E, 100×), scale bar, 100 μm. (B) Ultrastructural changes in the myocardium conducted by transmission electron microscopy (TEM, 10 000×); scale bar, 500 nm. MF: myofibrils; MI: mitochondria; Z: Z-line. Experimental groups are represented as follows: 1: control; 2: model; 3: Nat 1 mg/kg; 4: Nat 3 mg/kg; 5: Nat 9 mg/kg; 6: Gli 50 mg/kg; 7: Gli 50 mg/kg+Nat 3 mg/kg; 8: Ipt 3 mg/kg; 9: Lis 15 mg/kg.
Under TEM, myocardial tissues derived from the rats treated with ISO or ISO combined with Gli showed myocardial edema, irregular nuclei, Z line (Z) disorder, and mitochondrial (MI) swelling, with fuzzy ridges and myofilament dissolution. However, the MF and Z lines were orderly arranged with clear filaments, and the mitochondria were uniformly dispersed with clear ridges and uniform nuclear chromosomes in the ISO-CHF rats treated with Nat or Ipt. The changes in the myocardial ultrastructure could also be improved by treatment with Nat or Ipt (Figure 2B). Nat and Ipt had similar and equivalent effects at equal doses, and Nat effects could be prevented by treatment with Gli in vivo.
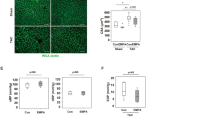
As shown in Figure 3A, the yellow and dark yellow colors represent the alpha smooth muscle actin (α-SMA) expression in the different groups. To measure the α-SMA expression levels, we analyzed the integrated optical density (IOD) using a microscope (Figure 3B). The α-SMA expression was significantly upregulated in the ISO-CHF rats (P<0.01). Nat, Ipt, and Lis could reverse the overexpression of α-SMA induced by ISO, but Nat effects could be antagonized by Gli in vivo.
Effects of natakalim on alpha-smooth muscle actin (α-SMA) levels in ISO-CHF rats. (A) Immunohistological staining of α-SMA (100×); scale bar, 100 μm. Yellow and dark yellow colors indicate α-SMA expression. α-SMA is one of the important indices of the degree of myocardial fibrosis. (B) Microscopic analysis of α-SMA expression. The data are expressed as the mean±SD. n=3. **P<0.01 vs control. ##P<0.01 vs model. $$P<0.01 vs Nat 3 mg·kg−1·d−1. Integrated optical density (IOD) represents α-SMA expression. Experimental groups are as follows: 1: control; 2: model; 3: Nat 1 mg/kg; 4: Nat 3 mg/kg; 5: Nat 9 mg/kg; 6: Gli 50 mg/kg; 7: Gli 50 mg/kg+Nat 3 mg/kg; 8: Ipt 3 mg/kg; 9: Lis 15 mg/kg.
To observe the degree of cardiac hypertrophy, we measured the heart size and determined the heart weight/body weight ratio (HW/BW, Figure 4). As shown in Figure 4A and 4B, the HW/BW and heart size significantly increased in the ISO group compared to that in the control group. Both Nat and Ipt could decrease heart size and HW/BW. Nat effects could be antagonized by Gli in vivo.
Cardiac hypertrophy improvement. (A) Heart size. The scale bar is the same. Experimental groups are represented as follows: 1: control; 2: model; 3: Nat 1 mg/kg; 4: Nat 3 mg/kg; 5: Nat 9 mg/kg; 6: Gli 50 mg/kg; 7: Gli 50 mg/kg+Nat 3 mg/kg; 8: Ipt 3 mg/kg; 9: Lis 15 mg/kg. (B) HW/BW. The data are expressed as the mean±SD. n=10. **P<0.01 vs control. ##P<0.01 vs model. $$P<0.01 vs Nat 3 mg/kg.
CHF biomarkers
As illustrated in Figure 5A and 5B, the plasma levels of NT-proBNP increased (P<0.01), and the BNP mRNAs were overexpressed in the ISO-CHF rats. However, these changes were completely restored to normal levels following treatment with Nat for 8 weeks at all the tested doses. At the same dosage (3 mg), Nat and Ipt exhibited equivalent effects, while those of Nat could be prevented by Gli in vivo.
Effects of natakalim (Nat) on N-terminal prohormone brain natriuretic peptide (NT-proBNP) and BNP levels in the ISO-CHF rats. (A) Plasma levels of NT-proBNP. (B) BNP mRNA expression in hearts. All data are expressed as the mean±SD. n=6. **P<0.01 vs control. ##P<0.01 vs model. $$P<0.01 vs Nat 3 mg/kg.
Endothelial dysfunction improvement
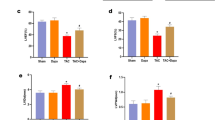
As shown in Figure 6A–6C, an increase in the serum levels of NO was observed along with a simultaneous decrease in the mRNA expression of eNOS. The mRNA expression of iNOS increased in the cardiac tissues derived from the ISO-CHF rats. These changes in the NO system could be improved by Nat and Ipt, but not by Lis. In addition, both the plasma levels of ET-1 and the levels of ET-1 mRNA expression in the cardiac tissues significantly increased in the ISO-CHF rats. This increase could be reversed by treatment with Nat for 8 weeks at all the doses tested (Figure 6D, 6E). At the same dosage (3 mg), Nat and Ipt showed equivalent effects, while Lis also significantly decreased the levels of ET-1. Nat effects could be blocked by Gli in vivo (Figure 6).
Effects of natakalim on nitric oxide (NO) and endothelin-1 (ET-1) levels in ISO-CHF rats. (A) Serum levels of NO. n=6. (B, C) eNOS and iNOS mRNA expression in hearts. n=6. (D) Plasma levels of ET-1. n=10. (E) ET-1 mRNA expression in hearts. n=6. All data are expressed as the mean±SD. **P<0.01 vs control. #P<0.05, ##P<0.01 vs model. $$P<0.01 vs Nat 3 mg/kg.
Analysis of anti-CHF mechanisms mediated by the SUR2B/Kir6.1 channel
Differential protein expression
The total plasma proteins were extracted from the control, model, and Nat (3 mg) groups in two and three independent biological experiments, and the protein profiles were analyzed using iTRAQ. After merging the data, 258 227 spectra were generated, of which 31 023 spectra matched known peptides, and 28 368 spectra matched unique peptides. Ultimately, 4043 peptides, 3578 unique peptides, and 724 proteins were identified. The GO and COG analysis annotated 724 proteins in different cellular components. Their molecular function and biological processes are described in the supplemental information (Supplementary Figure S1A–S1C and S2).
The selection criteria were as follows: significant iTRAQ alteration ratio for a particular protein and a minimum unused score of >1.2 or a maximum unused score of <0.8-fold (which indicates >95% confidence in correct protein sequence identification) by biological comparison between two groups. Fifty-five proteins were identified between the model and Nat (3 mg) treatment group and selected for further analysis using bioinformatic tools, and 33 and 22 of these proteins were upregulated and significantly downregulated, respectively (P<0.05, Supplementary Table S1). There was a significant difference in 66 proteins between the control and the model groups (Supplementary Table S2).
Functional enrichment analysis
An enrichment analysis of the differentially expressed proteins was conducted. The accumulated proteins were classified into three groups: biological process, cellular component, and molecular function. Among the 55 differential proteins, 42 accumulated proteins were annotated to certain GO categories, including biological process and molecular function categories. The annotated proteins were strongly enriched in the biological process categories: single-organism process (P=0.0179), cell communication (P=0.0179), signal transduction (P=0.0342), cellular responses to stress (P=0.0345), oxidation-reduction process (P=0.0326), ATP catabolic process (P=0.0024), endothelium development (P=0.0230), cardiac chamber development (P=0.0230), and endothelial cell differentiation (P=0.0230). The 42 annotated proteins were also significantly enriched in molecular functional properties (Table 4): ATPase activity (P=0.0125), iron ion binding (P=0.0250), retinol binding (P= 0.0045), proton-transporting ATPase activity, and rotational mechanism (P=0.0129). The GO analysis showed that Nat improved CHF through the candidate proteins involved in the above functions, including the proteins involved in ATP catabolic process.
Pathway analysis
Based on pathway analysis, Nat distinctly affected the following key pathways closely related to CHF. Many differentially accumulated proteins were further investigated using the KEGG database and were found to be mainly enriched (Table 5) in dilated cardiomyopathy (10.2%), viral myocarditis (12.24%), metabolic pathways (10.2%), oxidative phosphorylation (4.08%), calcium signaling pathway (10.2%), NF-kappa B signaling pathway (10.2%), mitogen-activated protein kinase (MAPK) signaling pathway (4.08%), leukocyte trans-endothelial migration (6.12%), apoptosis (2.04%), and the renin-angiotensin system (RAS, 2.04%). The candidate target proteins were accumulated in different pathways highly associated with CHF (Table 5). These results indicate that these key signaling pathways might play a pivotal role in the effect of Nat against CHF through the SUR2B/Kir6.1 channel.
Protein-protein interaction networks
To further understand the interaction spectrum of identified proteins, protein-protein interaction networks among the differential proteins were generated using STRING 9.0. A highly connected network composed of 55 proteins was successfully mapped (Figure 7). The majority of the identified interacting proteins fell into four clusters: proteasome (Psm) system function, ATP energy metabolic processing, stress, and the cAMP-dependent PKA signaling process. Three functional modules were apparent on this network, and they formed tightly connected clusters. The first functional module included Psm alpha 1 subunit (Psma1), Psma2, Psma5, Psmb1, Psmb2, Psmb4, Psmb5, Psmb6, and Psmc2. The second module included proteins involved in a large number of interactions in ATP energy metabolic processing, such as ATP5A1, ATP5B, ATP5C1, ATP5D, and ATP5O. The third module included HSP90b1, HSPa5, and Rbp4, which were associated with responses to stress. Numerous connectivities were to PRKAR2β, GAS6, Rap1b, AGT, and Tgfb1i1. This network map illustrated the improvement caused by Nat in CHF, which was mediated by the differential proteins involved in one of the functional groups or the whole interaction network.
Association network of proteins closely related to natakalim. (A) Protein metabolism: Psma1, Psma2, Psma5, Psmb1, Psmb2, Psmb4, Psmb5, Psmb6, Psmc2, and Rps27a. (B) Energy metabolism: ATP5A1, ATP5B, ATP5C1, ATP5D, and ATP5O. (C) Stress: HSP90b1, HSPa5, and Rbp4. (D) Signaling transduction: Prkar2β, Gas6, Rap1b, AGT, and Tgfb1i1. This network map illustrates that the different proteins involved in functional groups or the whole interaction network played a vital role in improving the effect of Nat against CHF. Proteins and their interactions are shown as nodes and edges. Proteins without connections are not included.
Important protein validation
To validate the iTRAQ data, we selected and measured TIMP3, AGT, ATP, PRKAR2β, VASP, and GAS6 by ELISA. The reasons for choosing them were as follows. (i) They were the most significantly expressed proteins in our list (P<0.05, Supplementary Table S2). (ii) According to the informational analysis, these proteins might participate in the biological processes and pathways related to the effect of Nat against CHF, which might indicate new mechanisms for CHF treatment. (iii) They might provide new insights into Nat′s molecular mechanism. The changes in these proteins showed significant differences among the control, ISO-CHF, and Nat treatment groups. The ELISA confirmed that the levels of TIMP3, ATP, PRKAR2β, VASP, and GAS6 were significantly lower in the model group than in the control (P<0.05), and the level of AGT was significantly higher than that in the control group (P<0.05) (Figure 8). After treatment, we observed a significant elevation in the plasma levels of TIMP3, ATP, PRKAR2β, VASP, and GAS6 (P<0.05). On the contrary, the level of AGT was significantly decreased (P<0.05). The concentrations of these proteins are shown in the scatter plots (Figure 8A–8F).
Effects of natakalim on the plasma levels of TIMP3, AGT, ATP, GAS6, PRKAR2β, and VASP. The data are expressed as individual levels. The black line represents the mean of individual level and standard deviation (SD). n=9. P-values are represented to have statistical significance between two groups: (A) **P=0.002 vs control and ##P=0.0011 vs model. (B) *P=0.0198 vs control and ##P=0.0025 vs model. (C) *P=0.0255 vs control and ##P=0.0090 vs model. (D) *P=0.0226 vs control and #P=0.0102 vs model. (E) *P=0.0176 vs control and ##P=0.0068 vs model. (F) *P=0.0230 vs control and ##P=0.0012 vs model.
Validation of eNOS/VASP pathway in vitro
As illustrated in Figure 9A, Nat could increase the NO content in cell culture medium, compared to that in the control. In contrast, the level of NO decreased in the NOS inhibitor L-NAME group and the L-NAME combined with Nat group. The Western blot results (Figure 9B) showed that Nat could significantly upregulate VASP expression, while L-NAME could downregulate it. The expression of VASP in the group treated with Nat combined with L-NAME did not significantly increase compared to that in the L-NAME-treated group.
Validation of eNOS/VASP pathway in EA.hy926 cell line. (A) NO content in the culture medium. (B) VASP expression in HUVECs. The data are expressed as the mean±SD. n=3. **P<0.01 vs control. ##P<0.01 vs Nat.
Discussion
Pharmacological validation for the SUR2B/Kir6.1 channel pathway against CHF
Equivalent effects of the two SUR2B/Kir6.1 channel openers against CHF
In this study, we compared the therapeutic effects of Nat and Ipt, at the same dose of 3 mg/kg and in the same ISO-CHF experiment. The results indicated that Nat and Ipt showed equivalent effects against ISO-CHF. In our previous study, the effects of Ipt were observed in PO-CHF at doses of 1, 3, and 9 mg/kg, and Nat effect was observed in the PO-CHF and MI-CHF experiments. These results showed that both Ipt and Nat could significantly improve the hemodynamics, cardiac disorders, and cardiac remodeling, and could decrease the levels of atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP)7,8,9,10. In the present study, we observed the effects of Nat at doses of 1, 3, and 9 mg/kg, and the results were in accordance with previously obtained results7,8,9,10. Therefore, these results demonstrated that the two SUR2B/Kir6.1 channel openers showed equivalent effects against CHF in the same experiment and that the drugs with different chemical structures aimed at the same target had similar effects in the same HF model. The results are of great value for the development of such HF drugs.
Antagonism of the KCB, Gli, against the SUR2B/Kir6.1 channel opener, Nat, in vivo
SUR2B/Kir6.1 channels are mainly distributed in blood vessels and are closely associated with endothelial function6,14,15. Our series of experiments using electrophysiological and molecular biological methods confirmed that both Nat and Ipt could open the SUR2B/Kir6.1 channels present on the endothelial cells with high selectivity16. In the present study, we chose the highly selective KCB, Gli, and combined it with Nat in vivo. The results showed that the effects of Nat against CHF could be blocked by Gli, confirming that the activation of the SUR2B/Kir6.1 channel is an effective therapeutic pathway for Nat against CHF. These results provide vital experimental proof that the actions of this agent against CHF are mediated by the SUR2B/Kir6.1 channel.
Differences in pharmacological mechanisms between the SUR2B/Kir6.1 channel openers and the ACEI, Lis
Both Nat and Ipt could significantly reduce the increased NO serum levels and overexpressed iNOS mRNA levels. However, they increased the protective eNOS mRNA expression in the endothelial system, and therefore increased the release of NO from endothelial cells, which increased the blood supply to damaged areas of the heart. Several studies have reported that eNOS could improve cardiac remodeling17,18, which supports our present results. However, ET-1 enhances cardiac remodeling and promotes the development of HF19. We found that both Nat and Ipt could reduce ET-1 levels in the plasma and myocardial tissue. Thus, it was reasonable to conclude that both Nat and Ipt could adjust the balance between ET-1 and NO systems. The results of this study were in accordance with the conclusion because they confirmed the modulatory effects of Nat and Ipt on the balance between the ET-1 and NO systems. In addition, the results of this experiment showed that the ACEI, Lis, could improve CHF, but through a completely different mechanism. Our results indicated that Lis could regulate the ET system but had only a minor effect on the NO system, suggesting that the effects of Lis against CHF were not mediated through the endothelial system.
Molecular pathway for SUR2B/Kir6.1 channel opening against CHF
To further elucidate the molecular mechanisms underlying the activation of the SUR2B/Kir6.1 channel against CHF, we used proteomic methods to study the changes in all the plasma proteins. From all the plasma protein changes, 724 proteins were identified, of which 55 proteins were expressed differentially (P<0.05) after Nat 3 mg treatment. The GO analysis revealed the biological processes and molecular functions associated with CHF (Table 4), including ATP catabolic process, endothelium development, and cardiac chamber development. The interaction analysis also provided a complete picture of the biological functions of the differential proteins, suggesting they have multiple roles among biological processes related to Psm, energy metabolism, stress, and signal transduction. Several studies indicated that ISO induces the stress response in the heart, which leads to relative myocardial ischemia and hypoxia20,21. Our results also demonstrated that Nat could restore the levels of ATP in the plasma of ISO-CHF rats (Figure 8F). Based on this evidence, we suggested that Nat could regulate the expression of PKA/HSP through the SUR2B/Kir6.1 channel to improve energy metabolism, restore Psm function, and ultimately reverse the structural changes in the myocardium. The results were also supported by histological and TEM analyses (Figure 2A, 2B). It is reasonable to propose that the interacting proteins in the sub-clusters of Psm, ATP, HSP, and PKA (Figure 7) were involved in the effects against CHF.
Furthermore, additional pathway analyses were carried out to prove that the differentially expressed proteins were involved in the key signaling pathways classified into 6 categories (Table 5), including energy metabolism, as well as cell growth and development. It has been reported that the metabolism of substances and energy intake of myocardial cells are affected by ISO-CHF22. This study showed that Nat could improve the energy metabolism of the heart (Figure 8F). In addition, our previous studies revealed that Nat could increase intracellular K+ outflow, trigger Ca2+ inflow in endothelial cells, increase eNOS mRNA expression, and thereby increase endothelial NO release23,24. Consistent with these findings, the targeting of the SUR2B/Kir6.1 channel in endothelial cells by Nat could regulate blood vessel tension through a series of intracellular signal transduction pathways, which play an important role in the regulation of blood vessels. Furthermore, this could affect cell development and function in blood vessels, and thereby correct endothelial dysfunction. Therefore, it was reasonable to speculate that the putative target proteins involved in these pathways were associated with the subsequent molecular mechanisms through the activation of the SUR2B/Kir6.1 channel.
Based on the function annotation and pathway analysis, we selected 6 candidate proteins closely related to the effects of Nat against CHF, and the ELISA results further proved that the iTRAQ data were reliable. In the present study, PRKAR2β, GAS6, and VASP were consistently upregulated after Nat treatment. Our previous results showed that Ipt could enhance the activity of PKA through the SUR2B/Kir6.1 channel in smooth muscle cells, and decrease the intracellular Ca2+ concentration, thereby regulating blood vessel tension25. In addition, another study proved the relationship between the PKA pathway and HF progression26. Thus, we can conclude that Nat increased the plasma levels of PRKAR2β, activated PKA, and decreased the intracellular Ca2+ concentration in smooth muscle cells. In this case, the blood vessels were relaxed, and the blood flow was increased, which could improve myocardial ischemia to achieve CHF improvement.
GAS6 is essential for stable cell-cell conjunctions27, and has been reported to regulate the phosphoinositide 3-kinase (PI3K)/Akt pathway and produce NO through the PI3K/Akt/eNOS pathway in endothelial cells28,29,30. Our results showed that GAS6 was elevated in the Nat treatment group, suggesting that it could participate in regulating NO through the PI3K/Akt/eNOS pathway in endothelial cells. VASP is involved in maintaining endothelial function in the blood vessels through the NO/cyclic guanosine monophosphate (cGMP)/protein kinase G (PKG) signaling pathway; VASP could protect the function of endothelial cells in vivo during hypoxia31,32,33.
These published data supported our findings that Nat could exert an effect against CHF by protecting the endothelial function. Taken together, these results suggested that Nat could upregulate PRKAR2β through the SUR2B/Kir6.1 channel and promote NO production through the GAS6/eNOS pathway in the endothelial cells. Furthermore, NO could activate cGMP-dependent protein kinase (PKG). Finally, VASP, as a downstream molecule regulated by PKG, had a vital effect on vascular endothelial function (Figure 9). Therefore, it is reasonable to suggest that Nat could protect endothelial function via PRKAR2β and the pathways related to NO, which provided novel insights into the molecular mechanism against CHF. To validate the relation between NO and VASP, we investigated VASP expression in HUVECs treated with Nat and the NOS specific anti-agent, L-NAME. These results demonstrated that Nat could improve endothelial function through the eNOS/VASP pathway, thereby achieving its therapeutic effect against CHF.
The appropriate expression of matrix metalloproteinases (MMPs) and the balance of MMPs/TIMPs have been suggested as vital factors for maintaining myocardial collagen fibers and cardiac structure34,35. We found that TIMP3 was significantly elevated after Nat treatment, which is beneficial for resolving CHF. The myocardial interstitial fibrosis was induced by the activation of the RAS in both the circulatory system and local tissues36. Our pathway analysis results also showed that AGT was positively correlated with the RAS. Therefore, we had reasons to confirm that the change in plasma levels of TIMP3 and AGT resulted from biological effects that were mediated by the SUR2B/Kir6.1 channel opening.
Conclusion
These results indicate that both SUR2B/Kir6.1 channel openers, Nat and Ipt, have equivalent pharmacological effects in improving endothelial dysfunction by rebalancing the NO and ET systems. The antagonistic effects of a highly selective KCB, Gli, in vivo demonstrated a new therapeutic pathway against CHF through the SUR2B/Kir6.1 channel in endothelial cells. In this experiment, we first identified 55 proteins closely related to Nat. These differentially expressed proteins were mainly involved in single-organism processes and energy metabolism, and we screened out PRKAR2β, GAS6/eNOS/NO and NO/PKG/VASP pathways involved in the amelioration of CHF among the 24 enriched pathways. We further confirmed 6 protein candidates, including PRKAR2β, GAS6, and VASP, involved in the endothelial mechanisms, and TIMP3, AGT, and ATP, which contribute to the cardiovascular actions. The Western blot results in the EA.hy926 cells demonstrated that the eNOS/VASP pathway was involved in its pharmacological mechanisms. Therefore, it is reasonable to suggest that the eNOS/VASP pathway is closely related to these newly identified therapeutic pathways through the SUR2B/Kir6.1 channel (Figure 10) in the endothelial cells. Finally, our findings provide new insights into the anti-CHF mechanism mediated through the SUR2B/Kir6.1 channel.
Summarized anti-CHF pathways mediated through the SUR2B/Kir6.1 channel. Both SUR2B/Kir6.1 channel openers, Nat and Ipt, confer protection of vascular function against CHF in the ISO-treated rats via the SUR2B/Kir6.1 channel opening that corrects the imbalance between NO and ET-1 systems. Mechanisms include activating the SUR2B/Kir6.1 channel in the endothelial cells, increasing GAS6 release, up-regulating PRKAR2β and VASP expression, restoring endothelial function, and improving CHF.
Author contribution
Hai WANG designed the research; Shang WANG conducted all the experiments; Chao-liang LONG, Jun CHEN, Yan-fang ZHANG, Wen-yu CUI, and Hao ZHANG performed parts of the experiments; and Shang WANG analyzed the data and wrote the paper.
References
Jessup M, Brozena S . Heart failure. N Engl J Med 2003; 348: 2007–18.
Ryan PM, Lawrence C, Shah PK . Chronic heart failure. Am J Cardiovas Drugs 2011; 11: 153–71.
Packer M, Carver JR, Rodeheffer RJ, Ivanhoe RJ, DiBianco R, Zeldis SM, et al. Effect of oral milrinone on mortality in severe chronic heart failure. N Engl J Med 1991; 325: 1468–75.
Farquharson CA1, Butler R, Hill A, Belch JJ, Struthers AD . Allopurinol improves endothelial dysfunction in chronic heart failure. Circulation 2002; 106: 221–6.
Tang X, Luo YX, Chen HZ, Liu DP . Mitochondria, endothelial cell function, and vascular diseases. Front Physiol 2014; 5: 175.
Wang H, Long CL, Duan ZB, Shi CG, Jia GD, Zhang YL . A new ATP-sensitive potassium channel opener protects endothelial function in cultured aortic endothelial cells. Cardiovasc Res 2007; 73: 497–503.
Gao S, Long CL, Wang RH, Wang H . KATP activation prevents progression of cardiac hypertrophy to failure induced by pressure overload via protecting endothelial function. Cardiovasc Res 2009; 83: 444–56.
Tang Y, Long CL, Wang RH, Cui WY, Wang H . Activation of SUR2B/Kir6.1 subtype of adenosine triphosphate-sensitive potassium channel improves pressure overload-induced cardiac remodeling via protecting endothelial function. J Cardiovasc Pharmacol 2010; 56: 345–53.
Zhou HM, Zhong ML, Zhang YF, Cui WY, Long CL, Wang H . Natakalim improves post-infarction left ventricular remodeling by restoring the coordinated balance between endothelial function and cardiac hypertrophy. Inter J Mol Med 2014; 34: 1209–18.
Zhong ML, Zhou HM, Long CL, Zhang YF, Cui WY, Wang H . Natakalim ameliorates isoproterenol-induced chronic heart failure by protecting against endothelial dysfunction. Pharmacology 2016; 98: 99–110.
Stéphanie FB, William CB, Adam AD, David ES, Peter CC, Joseph DT, et al. Plasma membrane proteomes of differentially matured dendritic cells identified by LC-MS/MS combined with iTRAQ labelling. J Proteomics 2012; 75: 938–48.
James DB, John PR, Arunangshu D, Jason L, Todd MU, Anne S, et al. Proteomic profiling of human plasma by iTRAQ reveals down-regulation of ITI-HC3 and VDBP by cigarette smoking. J Proteome Res 2011; 10: 1151–9.
Uros R, Kjell P, Jaco CK, Maarten L, Se bastien B, Oleg K, et al. iTRAQ-based proteomics profiling reveals increased metabolic activity and cellular cross-talk in angiogenic compared with invasive glioblastoma phenotype. Mol Cell Proteomics 2009; 8: 2595–612.
Stephan D, Winkler M, Kühner P, Russ U, Quast U . Selectivity of repaglinide and glibenclamide for the pancreatic over the cardiovascular K(ATP) channels. Diabetologia 2006; 49: 2039–48.
Seino S, Miki T . Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog Biophys Mol Biol 2003; 81: 133–76.
Wang SY, Cui WY, Wang H . The new antihypertensive drug iptakalim activates ATP-sensitive potassium channels in the endothelium of resistance blood vessels. Acta Pharmacol Sin 2015; 36: 1444–50.
Scherrer CM, Ullrich R, Bloch KD, Nakajima H, Nasseri B, Aretz HT, et al. Endothelial nitric oxide synthase limits left ventricular remodeling after myocardial infarction in mice. Circulation 2001; 104: 1286–91.
Kobayashi N, Mori Y, Nakano S, Tsubokou Y, Shirataki H, Matsuoka H . Celiprolol stimulates endothelial nitric oxide synthase expression and improves myocardial remodeling in deoxycorticosterone acetate-salt hypertensive rats. J Hypertens 2001; 19: 795–801.
Bras-Silva C, Castro-Chaves PM, Fontes-Sousa AP, Nunes P, Monteiro-Sousa D, Duarte AJ, et al. Impaired systolic and diastolic myocardial response to ET-1 and Ang II in heart failure. Eur J Heart Fail 2006; 5: S56–7.
Liu TT, Le CN, Kime EJ, Trand D, Phinney BS, Anne AK . Mitochondrial proteome remodeling in ischemic heart failure. Life Sci 2014; 101: 27–36.
Planavila A, Redondo AI, Ribas F, Garrabou G, Casademont J, Giralt M, et al. Fibroblast growth factor 21 protects the heart from oxidative stress. Cardiovasc Res 2015; 106: 1–13.
Tuunanen H, Knuuti J . Metabolic remodeling in human heart failure. Cardiovasc Res 2011; 90: 251–7.
Chen XJ, Han WZ, Zhang YF, Cui WY, Pan ZY, Long CL, et al. The molecular pathway of ATP-sensitive potassium channel in endothelial cells for mediating arteriole relaxation. Life Sci 2015; 137: 164–9.
Zhao Fl, Fu L, Yang W, Dong YH, Yang J, Sun SB . Cardioprotective effects of baicalein on heart failure via modulation of Ca2+ handling proteins in vivo and in vitro. Life Sci 2016; 145: 213–23.
Gao M, Wang Y, Wang H . Effects of iptakalim on intracellular calcium concentrations, PKA and PKC activities in rat tail artery smooth muscle cells. Acta Pharmacol Sin 2005; 40: 954–7.
Wehrens XHT, Lehnart SE, Reiken S, Vest JA, Wronska A, Marks AR . Ryanodine receptor/calcium release channel PKA phosphorylation: a critical mediator of heart failure progression. Proc Natl Acad Sci U S A 2006; 103: 3511–8.
Hasanbasic I, Cuerquis J, Varnum B, Blostein MD . Intracellular signaling pathways involved in Gas6-Axl-mediated survival of endothelial cells. Am J Physiol Heart Circ Physiol 2004; 287: H1207–H1213.
Laurance S, Aghourain MN, Lila ZJ, Lemarie CA, Blostein MD . Gas6-induced tissue factor expression in endothelial cells is mediated through caveolin-1-enriched microdomains. J Thromb Haemost 2014; 12: 395–408.
Son BK, Kozaki K, Iijima K, Eto M, Nakano T, Akishita M, et al. Gas6/Axl-PI3K/Akt pathway plays a central role in the effect of statins on inorganic phosphate-induced calcification of vascular smooth muscle cells. Eur J Pharmacol 2007; 556: 1–8.
Wang B, Yang Q, Bai WW, Xing YF, Lu XT, Sun YY, et al. Tongxinluo protects against pressure overload-induced heart failure in mice involving VEGF/Akt/eNOS pathway activation. PloS One 2014; 9: e98047.
Furman C, Sieminski AL, Kwiatkowski AV, Rubinson DA, Vasile E, Bronson RT, et al. Ena/VASP is required for endothelial barrier function in vivo. J Cell Biol 2007; 179: 761–75.
Ibarra-Alvarado C, Galle J, Melichar VO, Mameghani A, Schmidt HH . Phosphorylation of blood vessel vasodilator-stimulated phosphoprotein at serine 239 as a functional biochemical marker of endothelial nitric oxide/cyclic GMP signaling. Mol Pharmacol 2002; 61: 312–9.
Schmit MA, Mirakaj V, Stangassinger M, König K, Köhler D, Rosenberger P . Vasodilator phosphostimulated protein (VASP) protects endothelial barrier function during hypoxia. Inflammation 2012; 35: 566–73.
Lindsey ML, Zamilpa R . Temporal and spatial expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases following myocardial infarction. Cardiovasc Ther 2012; 30: 31–41.
Kassiri Z, Oudit GY, Sanchez O, Dawood F, Mohammed FF, Nuttall RK, et al. Combination of tumor necrosis factor-α ablation and matrix metalloproteinase inhibition prevents heart failure after pressure overload in tissue inhibitor of metalloproteinase-3 knock-out mice. Circ Res 2005; 97: 38090.
Moria J, Zhang LY, Oudit GY, Lopaschuk GD . Impact of the renin-angiotensin system on cardiac energy metabolism in heart failure. J Mol Cell Cardiol 2013; 63: 98–106.
Acknowledgements
This work was supported by grants from the National Basic Research “973” Program (Grants No 2012CB518200 and JCKY2013000B001) and the State Key Research Project of China (Grant No AWS11J003).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary information is available on Acta Pharmacologica Sinica's web site.
Supplementary information
Supplementary Figure S1
Gene Ontology (GO) functional annotations of all proteins. (DOC 103 kb)
Supplementary Figure S2
Cluster of Orthologous Groups (COG) of all proteins. (DOC 82 kb)
Supplementary Table S1
The differential proteins in the plasma of model and Nat 3mg/kg treated group from three independent biological replicate experiments (DOC 90 kb)
Supplementary Table S2
The differential proteins in the plasma of control and model group from three independent biological replicate experiments (DOC 143 kb)
Rights and permissions
About this article
Cite this article
Wang, S., Long, Cl., Chen, J. et al. Pharmacological evidence: a new therapeutic approach to the treatment of chronic heart failure through SUR2B/Kir6.1 channel in endothelial cells. Acta Pharmacol Sin 38, 41–55 (2017). https://doi.org/10.1038/aps.2016.118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2016.118
Keywords
This article is cited by
-
The impact of oral anti-diabetic medications on heart failure: lessons learned from preclinical studies
Heart Failure Reviews (2018)