Abstract
Aim:
To investigate the action of isothiafludine (NZ-4), a derivative of bis-heterocycle tandem pairs from the natural product leucamide A, on the replication cycle of hepatitis B virus (HBV) in vitro and in vivo.
Methods:
HBV replication cycle was monitored in HepG2.2.15 cells using qPCR, qRT-PCR, and Southern and Northern blotting. HBV protein expression and capsid assembly were detected using Western blotting and native agarose gel electrophoresis analysis. The interaction of pregenomic RNA (pgRNA) and the core protein was investigated by RNA immunoprecipitation. To evaluate the anti-HBV effect of NZ-4 in vivo, DHBV-infected ducks were orally administered NZ-4 (25, 50 or 100 mg·kg−1·d−1) for 15 d.
Results:
NZ-4 suppressed intracellular HBV replication in HepG2.2.15 cells with an IC50 value of 1.33 μmol/L, whereas the compound inhibited the cell viability with an IC50 value of 50.4 μmol/L. Furthermore, NZ-4 was active against the replication of various drug-resistant HBV mutants, including 3TC/ETV-dual-resistant and ADV-resistant HBV mutants. NZ-4 (5, 10, 20 μmol/L) concentration-dependently reduced the encapsidated HBV pgRNA, resulting in the assembly of replication-deficient capsids in HepG2.2.15 cells. Oral administration of NZ-4 dose-dependently inhibited DHBV DNA replication in the DHBV-infected ducks.
Conclusion:
NZ-4 inhibits HBV replication by interfering with the interaction between pgRNA and HBcAg in the capsid assembly process, thus increasing the replication-deficient HBV capsids. Such mechanism of action might provide a new therapeutic strategy to combat HBV infection.
Similar content being viewed by others
Introduction
Although a hepatitis B vaccine has been available for more than 20 years, the estimated 400 million persons are still chronically infected with the hepatitis B virus (HBV) worldwide1. The currently approved antiviral therapies for hepatitis B virus infection include immunomodulators [interferon (IFN)-α and pegylated IFN-α)] and nucleoside or nucleotide analogues2. Because of poor tolerability, the adverse effects of IFN-α, and nucleotide analogue resistance of HBV, new potent antiviral agents are required for the improvement of chronic hepatitis B (CHB) treatment3. Thus, potential new targets, aside from HBV polymerase, have been explored and include viral RNA transcription factors, capsid formation, and envelope protein modification4. Because the HBV core protein (HBcAg) is highly conserved and the capsid provides the only site of HBV genome synthesis, the process of capsid assembly has been evaluated as a novel drug target for CHB therapeutics5.
To discover novel drugs that inhibit HBV replication, our high-throughput screening campaign yielded a series of structurally related, 2,2-bis-heterocycle tandem derivatives that potently inhibited HBV replication6. Here, we report a novel, potent HBV inhibitor, isothiafludine (NZ-4), which is a derivative of bis-heterocycle tandem pairs and is derived from the natural product leucamide A. Our results showed that as a small-molecule effector of HBV capsid assembly, NZ-4 was effective against the replication of HBV DNA by blocking the encapsidation of pregenomic RNA (pgRNA) and interfering in the assembly of nucleocapsids. Furthermore, NZ-4 efficiently inhibited the replication of HBV mutants that were resistant to nucleoside and nucleotide analogues. Hence, NZ-4 represented a novel chemical entity with a unique anti-HBV mechanism that warrants further development as a novel anti-HBV therapeutic for the treatment of chronic hepatitis B infection.
Materials and methods
Reagents
NZ-4, Bay38-76907, and 8–18 were synthesized in our institute. The structure of these compounds was identified using proton nuclear magnetic resonance, and the purity was determined as 99.5% using high-performance liquid chromatography (HPLC). 3TC was purchased from Tianfeng Chemical Technology Co, LTD (Suizhou, Hubei, China), and the purity was determined as 99.1% by HPLC.
Plasmids
The plasmid pHBV1.3 contains a 1.3-mer, over-length HBV genotype A2 genome (adw2 subtype; GenBank accession number: X02763.1)9 and was kindly provided by Prof Dr Dong-liang YANG (Tongji Hospital of Tongji Medical College, Wuhan, China). The HBV genome region that encoded the full-length Cp185 protein was amplified by PCR using pHBV1.3 as a template10,11. The plasmid expressing full-length HBcAg pHBc185 was constructed by cloning the respective PCR product into the vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). The plasmid pHBV1.3M204V/L180M, which contained a lamivudine/entecavir (3TC/ETV)-dual-resistant (rtM204V/rtL180M) HBV genome, and the plasmid pHBV1.3A181T/N236T, which contained an adefovir (ADV)-resistant (rtA181T/rtN236T) HBV genome, were generated by site-directed mutagenesis using a Site-directed Gene Mutagenesis Kit (Beyotime, Haimen, Jiangsu, China) according to instruction.
Cell culture and transient transfection
The HepG2.2.15 cell line is a HepG2 hepatoma-derived cell line that stably replicates HBV from four integrated, tandem copies of the HBV genome (ayw serotype, GenBank accession number: U95551)12 and was maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Scientific, Hyclone, USA) and 380 μg/mL G418 (Sigma-Aldrich Co LLC, St Louis, MO, USA). The Huh7 hepatoma cell line was grown in DMEM supplemented with 10% FBS. Huh7 cells were seeded in 6-well plates with 1×106 cells per well and transfected with 1 μg plasmid DNA using the Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. All cell lines were cultured at 37 °C in the presence of 5% CO2.
Cytotoxicity assay
Cell viability was determined using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Sigma-Aldrich Co LLC, St Louis, MO, USA) method. Briefly, HepG2.2.15 cells were seeded in 96-well culture plates at a density of 5×103 cells per well in triplicate with different concentrations of NZ-4. The culture medium was removed 4 d later and replaced with new medium that was supplemented with or without NZ-4. After 8 d of culture, the medium was removed from the cells, and 100 μL MTT (final concentration 2.5 mg/mL) reagent was added for 4 h at 37 °C. The cells were then lysed with 10% sodium dodecyl sulfate (SDS) and 50% N,N-dimethylformamide, pH 7.2. OD values were read at 570 nm, and the percentage of cell death was calculated13.
HBV DNA quantification by qPCR
HepG2.2.15 cells were cultured in 96-well plates at a density of 5×103 cells per well for 8 d under standard conditions. At d 4, the medium was changed, and new inhibitor was added. At d 8, HBV progeny DNA was extracted from the cell culture supernatants using the QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) and quantified using real-time quantitative polymerase chain reaction (qPCR), as described previously14. The sequences of the HBV-specific fluorogenic probe (HBVayw/Probe) and primers for HBV (HBVayw/Sp and HBVayw/Asp) were as follows: HBVayw/Sp, 5′-CACCTCTCTTTACGCGGACT-3′, HBVayw/Asp, 5′-CGACGTGCAGAGGTGAAG-3′, and HBVayw/Probe, 5′-ATCTGCCGGACCGTGTGCAC-3′.
HepG2.2.15 cells were cultured in 24-well plates at a density of 1×105 cells per well. After an 8-d treatment with different concentrations of NZ-4, the cells were collected, and the capsid-associated HBV DNAs from the intracellular HBV capsids were extracted. Briefly, the cells were lysed with cold lysis buffer (0.5% NP-40, 50 mmol/L Tris·HCl, 1 mmol/L EDTA·2 Na, pH 7.0) at 4 °C for 15 min. The cell lysate was then digested with 100 U/mL Cyronase cold-active nuclease (TaKaRa, Dalian, China) for 30 min at 37 °C and then incubated with proteinase K at a concentration of 500 mg/mL at 56 °C for 2 h to release the associated HBV DNA. Capsid-associated DNA was purified by phenol/chloroform (1:1, v/v) extraction and then precipitated in 3 vol of ethanol15. The nucleic acid pellets were resuspended in TE buffer and quantified using qPCR, as described above.
Southern blotting analysis
HepG2.2.15 cells were cultured in 24-well plates, and the HBV DNAs from the intracellular HBV capsids were extracted according to the protocol described above. HBV replicative intermediates were detected by Southern blotting with a modification that included using a DIG-labeled PCR fragment that encompassed genome position 463 to 149912. All given nucleotide positions began with the adenine of the initiation codon of the C gene. Briefly, HBV DNAs were separated by electrophoresis using 0.8% agarose and transferred to a nylon membrane (Hybond-N+ membrane, GE, Connecticut, USA) using the capillary transfer method with 20×SSC buffer overnight at room temperature. After UV crosslinking, the membrane was hybridized with the DIG-labeled HBV DNA probe overnight at 42 °C and washed with 2×SSC/0.1% SDS twice for 5 min at room temperature and 0.2×SSC/0.1% SDS twice for 20 min at 55 °C15. The membrane was immunoblotted with an antibody that recognized DIG (Roche, Mannheim, Germany) followed by detection using the DIG Luminescent Detection Kit (Roche, Mannheim, Germany) according to the manufacturer's protocol.
Native agarose gel electrophoresis analysis
After treatment with the compounds, the HepG2.2.15 cell lysate was prepared similarly and resolved on a 1.8% native agarose gel. The capsids were electrophoresed at 70 V with TAE buffer and then transferred directly to a nitrocellulose membrane (GE, Connecticut, USA) using the capillary transfer method with 10×SSC buffer overnight. The membranes were then immunoblotted with a primary antibody that recognized HBcAg (Abcam, Cambridge, UK) followed by detection with a secondary antibody using chemiluminescence. Another replicate was transferred to a nylon membrane (Hybond-N+ membrane, GE, Connecticut, USA) under the same conditions, but the encapsidated nucleic acid was released from the capsids by denaturation. Prehybridization and hybridization were performed identically to that for Southern blotting analysis16.
Western blotting analysis
The HepG2.2.15 cells that were treated with different concentrations of compounds were collected and lysed in SDS sample buffer (62.5 mmol/L Tris·HCl, pH 6.8; 2% SDS; 10% glycerol; 5% 2-mercaptoethanol; and 0.02% bromophenol blue) and boiled for 5 min at 100 °C. The samples were submitted to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with primary antibodies that recognized HBcAg (Abcam, Cambridge, UK) and actin (Abmam, Cambridge, UK). Subsequently, bound secondary antibodies were detected with a chemiluminescence substrate (GE, Connecticut, USA) and exposed to X-ray film (Sigma-Aldrich Co LLC, St Louis, MO, USA).
Immunofluorescence analysis
For immunofluorescence staining, HepG2.2.15 cells were cultured on non-coated, glass coverslips17. After an 8-d treatment with the compounds, the cells were rinsed with PBS twice, fixed with 4% paraformaldehyde in PBS for 10 min at room temperature, and blocked with 10% BSA in PBS for 1 h. The primary antibody that recognized HBcAg (Abcam, Cambridge, UK) and the secondary antibody that was coupled to fluorescein isothiocyanate (Abcam, Cambridge, UK) were diluted in blocking buffer containing 0.1% (w/v) saponin. The nuclei were counterstained with 5 μg/mL DAPI. The coverslips were mounted on slips in mounting medium, and the specimens were viewed using a fluorescence microscope with a 40×objective (Olympus, Tokyo, Japan).
ELISA
The levels of HBsAg and HBeAg in the HepG2.2.15 cell culture were measured using specific-ELISA kits (Sino-American Biotechnology Company, Henan, China) following the manufacturer's recommendations.
HBV RNA quantification by qRT-PCR
HepG2.2.15 cells were cultured in 24-well plates as described above. The cell lysates were digested with 20 U/mL Cyronase cold-active nuclease (TaKaRa, Dalian, Liaoning, China) for 15 min at 4 °C, and encapsidated HBV pgRNA was isolated using the QIAamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany) that included digestion with a DNase Set (Qiagen, Hilden, Germany). Total RNA was extracted from HepG2.2.15 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The HBV RNA samples were quantified using qRT-PCR and the QuantiTect Virus Kit (Qiagen, Hilden, Germany). For each sample, qRT-PCR was performed in duplicate.
Northern blotting analysis
Total RNAs and the encapsidated pgRNA were isolated as described above. For Northern blotting, these samples were electrophoresed in a 1% formaldehyde agarose gel containing 5% formaldehyde and transferred to a nylon membrane (Hybond-N+ membrane, GE, Connecticut, USA). The transfer and hybridization were performed following the protocol of the NorthernMax® kit (Ambion, Austin, TX, USA).
RNA immunoprecipitation (RIP)
After a 48-h treatment with the compounds, pHBV1.3-transfected Huh 7 cells were treated with 37% formaldehyde for 20 min at room temperature to generate protein-RNA cross-links. After stopping the reaction with 2 mol/L glycine, a whole-cell extract was prepared in the presence of RNase inhibitors [50 mmol/L HEPES, pH 7.5; 140 mmol/L NaCl; 1 mmol/L EDTA; 1% (v/v) Triton X-100; 0.1% (w/v) sodium deoxycholate; and 40 U RNasin] to maintain the integrity of the RNA, and the extract was then treated with DNase I at a final concentration of 240 μg/mL to remove DNA. Immunoprecipitation was performed by incubating the lysates with rabbit polyclonal HBcAb (Abcam, Cambridge, UK) overnight at 4 °C, and protein A-Sepharose beads were added (equilibrated in lysis buffer containing 1 mg/mL BSA) for 1 h. The protein-RNA complexes were eluted from the beads with elution buffer (100 mmol/L Tris·HCl, pH 8.0; 10 mmol/L EDTA; 1% (w/v) SDS), and reversal of the formaldehyde cross-linking was performed by incubating the complexes at 65 °C. The immunoprecipitated pgRNA was detected using RT-PCR with a pair of HBV-specific primers that targeted the ɛ sequence at the 5′ end of the pgRNA.
Animals
3-d-old Hubei ducks (Tadorna tadorna) were obtained from a commercial hatchery (Chunjiang QinYe Limited Liability Company, Wuhan, China) and were held in the animal house of Tongji Hospital of Tongji Medical College, Huazhong University of Science and Technology. The ducklings were maintained under normal daylight and fed a standard, commercial diet, which was in accordance with the guidelines for animal care at the facilities of the Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences under permission of the Ethical Committee for Animal Experiments of the Institute of Medicinal Biotechnology, and given water. 3-d-old Hubei ducklings were experimentally infected via intra-peritoneal injection with the DHBV inoculation (1×107 copies/200 μL) (Day-10) that was collected from the supernatant of the culture transfected with the 1.5-fold-overlength genome recombinant plasmid18. 10 d after inoculation (Day 0), DHBV-positive ducklings were randomly divided into 6 groups that each contained 10 ducklings. The compounds were administered orally, twice per day for 15 d. Blood samples were obtained every 5 d, and serum samples from the experimentally infected ducklings were obtained at the initiation of treatment (Day 0), on the 5th day of treatment (Day 5), on the 10th day of treatment (Day 10), on the 15th d of treatment (Day 15), and on the 5th d (Day 20) of the post-treatment follow-up. The DHBV DNA in the sera was detected using qPCR with the sense primer (DHBV 1450F) 5′-GATACTGGAGCCCAAACC-3′ and anti-sense primer (DHBV 1737R) 5′-GGCAGAGGAGGAAGTCAT-3′.
Results
NZ-4 effectively inhibited HBV DNA replication in vitro
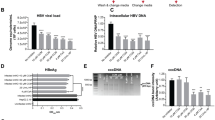
NZ-4 is a novel, non-nucleosidic anti-HBV compound, and its chemical structure is shown in Figure 1A. The cytotoxicity of NZ-4 was studied using MTT conversion assays in HepG2.2.15 cells (ayw serotype, wild type genome)14, and the concentration of 50% cytotoxicity (CC50) was 50.4 μmol/L after 8 d of culture (Figure 1B). In vitro, the anti-HBV activities of NZ-4 were examined using HepG2.2.15 cells, and qPCR was used to quantify the HBV DNA after treatment with NZ-4. The 50% inhibitory concentration (IC50) of NZ-4 for HBV DNA in HepG2.2.15 cell culture supernatants was 1.33 μmol/L (Figure 1C). The supernatant IC50 of the three nucleoside analogs, 3TC, ADV, and ETV, that were used in our assay are shown in Table 1 as a reference. The IC50 for intracellular HBV replication based on the detection of capsid-associated HBV DNA was 1.05 μmol/L (Figure 1D), which was much lower (≈50-fold) than the CC50 of HepG2.2.15 cells. These results confirmed the antiviral activity of NZ-4 against HBV.
NZ-4 efficiently inhibited HBV DNA replication in vitro. (A) Chemical structure of isothiafludine (NZ-4). (B) HepG2.2.15 cells were treated with NZ-4 at the indicated concentrations for 8 d. The cytotoxicity was determined using the MTT assay. The 50% cytotoxicity concentration (CC50) was calculated as the compound concentration that was necessary to reduce the cell viability by 50%. (C and D) HBV DNA in culture supernatants of HepG2.2.15 cells and capsid-associated HBV DNA in cells were quantified using qPCR. The 50% inhibitory concentration (IC50) was calculated as the concentration of the compounds that was required to reduce the HBV DNA load by 50%. The results are expressed as the mean±SD of three independent experiments.
Southern blotting analyses were performed to measure the HBV replication intermediates. The results showed that NZ-4 potently inhibited the various forms of the HBV intracellular DNA replication intermediates (RC, DSL, and SS HBV DNA) in a dose-dependent manner. 3TC was used as a positive control and inhibited HBV DNA synthesis effectively (Figure 2). Moreover, NZ-4 was active against various drug-resistant HBV mutants, including 3TC/ETV-dual-resistant and ADV-resistant HBV mutants, which indicated that the antiviral mechanism of NZ-4 is distinct from that of the nucleosidic inhibitors (Figure 3).
NZ-4 reduced the HBV replication intermediates with comparable efficiency. HepG2.2.15 cells were cultured in 6-well plates and treated with NZ-4 and/or 3TC at the indicated concentrations for 8 d. HBV replication intermediates were detected by Southern blotting hybridization using a DIG-labeled HBV genomic fragment as a probe. The various forms of the HBV replication intermediates were inhibited by NZ-4 in a concentration-dependent manner. RC, relaxed circular HBV DNA; DSL, double-stranded linear DNA; SS, single-stranded HBV DNA. As a polymerase inhibitor, 3TC specifically decreased the amount of intracellular HBV DNA.
NZ-4 inhibited the replication of 3TC/ETV-dual-resistant and ADV-resistant HBV mutants. The plasmids pHBV1.3 with wild type (A), 3TC/ETV-dual-resistant (rtM204V/rtL180M) (B), and ADV-resistant (rtA181T/rtN236T) HBV genomes (C) were separately transiently transfected into Huh7 cells. The cells were treated with the compounds at the indicated concentrations for 48 h. Intracellular HBV replication intermediates were analyzed by Southern blotting hybridization. When compared with wild type HBV, the drug-resistant HBV mutants showed resistance to 3TC, ADV, and ETV but were sensitive to NZ-4 treatment.
NZ-4 reduced encapsidated RNA with no influence on total HBV RNA synthesis
To explore whether the observed reduction of HBV DNA replication in HepG2.2.15 cells by NZ-4 was due to its impact on HBV RNA synthesis or pgRNA encapsidation, Northern blotting was performed to measure the intracellular HBV RNA and encapsidated pgRNA levels after treatment with NZ-4. As shown in Figure 4A, HBV RNA synthesis was not affected by NZ-4 or the negative control, 3TC, in HepG2.2.15 cells. As a positive control, the transcription inhibitor, 8–1, significantly inhibited the synthesis of the 3.5-kb and 2.1/2.4-kb RNAs of HBV8. The encapsidated pgRNA was also isolated after digestion with DNase. Northern blotting results showed that treatment with increasing amounts of NZ-4 induced a dose-dependent decrease in the levels of encapsidated HBV pgRNA (Figure 4B). The total intracellular HBV RNAs and encapsidated pgRNA were quantified using qRT-PCR (Figure 4C), and the reduction in encapsidated pgRNA but not total HBV RNAs indicated that NZ-4 might interfere in capsid assembly.
NZ-4 treatment reduced the encapsidation of pgRNA. HepG2.2.15 cells were treated with or without NZ-4 at the indicated concentrations for 8 d. (A) Total cellular RNA was extracted and subjected to Northern blotting hybridization to detect the HBV transcripts. As a transcription factor inhibitor, 8–1 decreased the HBV RNA level. (B) After digestion with DNase, the isolated, encapsidated pgRNA was electrophoresed in a 1% formaldehyde agarose gel and detected by Northern blotting hybridization. The encapsidated pgRNA was significantly reduced by NZ-4 treatment. (C) Total intracellular HBV RNAs and encapsidated pgRNA were quantified using qRT-PCR. 3TC and 8–1 were used as the negative and positive control, respectively. The results are expressed as the mean±SD of three independent experiments.
NZ-4 treatment led to the accumulation of replication-deficient capsids
The HBV core proteins (HBcAg) assemble into nucleocapsids5; therefore, the intracellular HBcAg levels after treatment with NZ-4 were measured using Western blotting analyses. As observed for the transcription of HBV RNA, HBcAg expression was not affected by NZ-4 or 3TC in HepG2.2.15 cells. However, Bay38-7690 reduced the steady-state level of HBcAg, as reported7 (Figure 5A). Furthermore, HBcAg distribution was not affected by NZ-4 or 3TC in HepG2.2.15 cells but was inhibited by the transcription inhibitor 8–1 (Figure 5B). In addition, NZ-4 treatment showed no influence on the secretion of HBsAg and HBeAg (Figure 5C).
NZ-4 treatment enhanced the production of replication-deficient HBV capsids but had no effect on the expression, distribution, or secretion of the HBV protein. HepG2 and HepG2.2.15 cells were cultured in 6-well plates with different concentrations of compounds for 8 d. (A) The cell lysates were loaded onto an SDS-PAGE gel to detect HBcAg by Western blotting with a rabbit polyclonal HBcAb. The expression of HBcAg was similar in NZ-4-treated and control HepG2.2.15 cells (upper panel). Actin protein was detected as the loading control (lower panel). (B) The intracellular distribution of HBcAg was visualized using immunofluorescence microscopy. After fixation, HepG2.2.15 cells were stained for HBcAg (green), and the nuclei were stained with DAPI (blue). (a) HepG2 cells were used as a negative control. (b) Untreated HepG2.2.15 cells were used as a positive control for HBcAg staining. (c) HepG2.2.15 cells were treated with NZ-4 at 20 μmol/L. NZ-4 inhibited HBV replication completely at this concentration but did not show a visible effect on the intracellular distribution of HBcAg. (d) HepG2.2.15 cells were treated with NZ-4 at 2.5 μmol/L. (e and f) HepG2.2.15 cells were treated with 3TC and 8–1, respectively. 3TC had no effect on the expression HBcAg, while 8–1 inhibited HBcAg expression. (C) Secreted HBsAg and HBeAg in the supernatant were analyzed using a commercial ELISA. NZ-4 did not show a significant influence on the production of HBsAg and HBeAg, even at a dose of 20 μmol/L, which was an effective concentration to inhibit HBV DNA replication. (D) The cell lysates were prepared and analyzed for HBV capsids by native agarose gel electrophoresis and immunoblotting with a rabbit polyclonal HBcAb (upper panel). Capsid-associated HBV DNA was detected by the transfer of HBV capsids onto a nylon sheet followed by Southern blotting hybridization upon disruption of the capsids in situ (lower panel).
Because NZ-4 had no significant effect on the expression and distribution of HBcAg, we further investigated whether a failure in capsid formation was the cause of the reduced HBV DNA replication. To investigate the effect of NZ-4 on HBV capsid assembly, HBV capsids from HepG2.2.15 cells exposed or not exposed to drug treatment were characterized using 1.8% native (non-reducing) agarose gel electrophoresis. As shown in Figure 5D, NZ-4 disturbed the assembly of the HBV capsid. In addition to the normal HBV capsid, a faster-migrating capsid band that contained no HBV DNA was the dominant band in the NZ-4-treated samples. This type of capsid was also present in untreated cells, but NZ-4 treatment led to the accumulation of this DNA-free capsid in a dose-dependent manner. In contrast, 3TC, which is a well-characterized inhibitor of HBV replication that is used for the treatment of chronic HBV infections, did not affect HBV capsid formation but efficiently inhibited HBV DNA synthesis, as expected. Bay38-7690, which is a capsid assembly inhibitor, effectively reduced the amount of viral capsids and core-associated DNA7. These results indicated that NZ-4 inhibited HBV DNA replication by interfering with capsid assembly but did not affect the expression or secretion of viral protein.
NZ-4 interfered with the binding of pgRNA and HBcAg and reduced encapsidated HBV pgRNA.
HBV nucleocapsid formation begins when the RNA pregenome, HBV polymerase and HBcAg dimers complex forms19. Therefore, we then determined whether the pgRNA encapsidation process was directly blocked by NZ-4 treatment. To study the interaction of the pgRNA with HBcAg during NZ-4 treatment, RIP was performed. The HBcAg and pgRNA complex was immunoprecipitated by an antibody to HBcAg (HBcAb). The crosslinks were then reversed, and the immunoprecipitated HBcAg was detected by Western blotting (Figure 6A). In parallel, the immunoprecipitated RNA samples were identified by PCR to prove that DNA was clearly removed (Figure 6B), and this was followed by RT-PCR with a pair of primers that targeted the ɛ sequence at the 5′ end of the pgRNA after digestion of the cross-linked HBcAg. The results showed that NZ-4 significantly interfered with the interaction between the pgRNA and HBcAg in the capsid assembly process (Figure 6C).
NZ-4 interfered with the interaction between the pgRNA and HBcAg. Huh7 cells were transfected with the plasmid pHBV1.3, which contained a replication-competent, over-length HBV genome, or the expression plasmid pHBc185, which had full-length HBcAg. The cells were treated or not treated with the compounds for 48 h. The HBcAg and pgRNA complex was immunoprecipitated using HBcAb, and after reversal of the formaldehyde cross-linking, the immunoprecipitated HBcAg was monitored using Western blotting analysis (A), and the immunoprecipitated pgRNA was detected by PCR (B) or RT-PCR (C).
NZ-4 suppressed DHBV DNA replication in a DHBV-infected duck model.
The inhibitory effect of NZ-4 on DHBV replication in vivo was analyzed in experimentally infected ducklings. In the first set of experiments, we observed the level of DHBV DNA in duck serum after treatment with different concentrations of NZ-4 and 3TC. When compared with the vehicle-treated group, qPCR analysis of the DHBV DNA in the sera indicated that DHBV DNA replication was suppressed by oral administration of NZ-4 in a dose-dependent manner on Day 15 after treatment (Figure 7). The reduction of the serum level of DHBV-DNA indicates that NZ-4 also showed its anti-DHBV efficacy in the DHBV-infected duck model.
NZ-4 inhibited DHBV replication in experimentally DHBV-infected ducklings in vivo. Ten days after intra-peritoneal injection with a DHBV inoculation, DHBV-positive ducklings were treated with the indicated compounds, as described in the Materials and methods section. At initiation of treatment (Day 0), on the 5th d of treatment (Day 5), on the 10th day of treatment (Day 10), on the 15th d of treatment (Day 15), and on the 5th day (Day 20) of post-treatment follow-up, DHBV DNA in the duck serum was detected by qPCR. Oral administration of NZ-4 for 15 d reduced the serum level of DHBV-DNA in DHBV-infected ducks.
Discussion
The present study analyzed a bis-heterocycle tandem derivative, NZ-4, which is a potent, non-nucleosidic inhibitor against HBV that blocks pgRNA encapsidation and interferes with HBV capsid assembly. The novel chemical structure of NZ-4 was derived from a natural marine product, leucamide A. In primary cell-based screening, we found that NZ-4 effectively reduced the HBV DNA in the supernatant and the replicative intermediates in the cytoplasm, and 3TC/ETV-dual-resistant and ADV-resistant HBV mutants were sensitive to it. Further experiments showed that although the amount of transcribed pgRNA and translated core protein was not altered, the level of encapsidated pgRNA was reduced and the level of abnormal capsid without HBV DNA accumulated with treatments of increasing concentrations of NZ-4. However, its precise mechanism must be further explored. From these results, we hypothesized that NZ-4 inhibited HBV replication by blocking encapsidation of the pgRNA and interfering in the nucleocapsid assembly process. Without an RNA template, the genome could not be synthesized. NZ-4-induced abnormal capsids were replicate-deficient and had decreased HBV DNA; therefore, the deficient capsids without nucleic acid were incompetent for replication, let alone viral infection.
The knowledge of the HBV life cycle has increased over the past decades, and this has led to the identification of several potential targets such as viral entry, cccDNA formation, capsid assembly, and viral morphogenesis4. The HBV capsid assembly has been proven to be a specific target of HBV inhibitors5. NZ-4 induces the formation of replication-deficient HBV capsids by interfering with the binding of the pgRNA and core protein. Replication-deficient HBV capsids formed in the presence of NZ-4 would unlikely be infectious because the majority are free of HBV DNA.
Previously, heteroaryldihydropyrimidines (HAPs) and phenylpropenamides have been reported to disturb capsid formation and maturation4,5. HAPs, including Bay 41-4109, Bay 38-7690, and Bay 39-5493, were first identified to prevent the formation of the HBV capsid20,21. These analogues inhibited HBV replication through binding within the assembly domain of HBcAg at a site that bridged elements of the secondary structure within a capsid-bound monomer and, thereby, misdirected capsid assembly7. HAPs bind at a sequence of the assembly domain that is involved in forming inter-dimer contacts, whereas DHBV-derived recombinant capsids show no binding7,20; therefore, HAPs did not inhibit DHBV replication in cultured cells or in a DHBV-infected duck model.
Although NZ-4 targeted capsid assembly, its mechanism of action is unique in that NZ-4 might target the nucleic acid binding domain of HBcAg. Because of the highly similar amino acid sequence between the HBV and DHBV core proteins, NZ-4 showed its antiviral efficacy in the DHBV-infected duck model by a substantial drop in viremia (Figure 7). Two other phenylpropenamide derivatives, AT-61 and AT-130, were reported to be assembly accelerators, which increased the rate of capsid assembly4. The phenylpropenamides trap HBV capsid assembly intermediates and leads to the formation of empty capsids but does not misdirect assembly22,23. The derivative AT-130 bound to the assembly domain of HBcAg with weak affinity and induced the protein into a more assembly active state. This increase in the concentration of the dimer in the active conformation resulted in an increased nucleation rate that drove capsid assembly forward22,23. In contrast, NZ-4 did not increase the rate of capsid assembly in vitro, and the total amount of intracellular HBV capsids was not affected by NZ-4. Moreover, these defective capsids showed a significantly different electrophoretic mobility after NZ-4 treatment (Figure 4A, 1st panel, lower band in each lane). We propose that the loss of intra-capsid components, such as HBV genomic material and/or proteins, will cause the HBV capsids to exert a different behavior by gel analysis, and NZ-4 strongly augments the formation of these replication-deficient capsids.
The HBV nucleocapsid that is formed by HBcAg is divided into N-terminal and C-terminal domains. The C-terminal domain is essential for packaging of the pgRNA/HBVPol complex, controls the interaction with HBV pgRNA, and has a strong influence on HBV DNA synthesis. We propose that the C-terminus of the core protein is more than a wielding hook for pgRNA attachment, and it might actively participate in regulating HBV DNA synthesis. To date, the topological structure of the C-terminal domain has not been determined by complete, core protein background crystallography due to its flexible nature. NZ-4 blocked the interaction between pgRNA and the core protein and reduced the encapsidation of the pgRNA.
This novel mechanism could provide a new perspective for the role of the HBV core protein C-terminal domain in HBV RNA encapsidation and HBV capsid assembly and might extend insights into the viral encapsidation of other related viruses. Taken together, the bis-heterocycle tandem derivative NZ-4 is shown to be a promising anti-HBV drug candidate. Its mechanism is different from other HBV inhibitors, and it is also found to effectively inhibit drug-resistant HBV mutants. The efficacy of NZ-4 on DHBV-infected ducklings warrants its further clinical investigation. Considering its specific mechanism of action, NZ-4 may provide a useful addition to anti-HBV drug research and development.
Author contribution
Li YANG, Li-ping SHI, and Hai-jun CHEN performed all experiments; Xian-kun TONG, Gui-feng WANG, Yang-ming ZHANG, Wen-long WANG, Chun-lan FENG, Pei-lan HE, and Feng-hua ZHU technically assisted in performing a portion of the experiments; You-hua HAO, Bao-ju WANG, and Dong-liang YANG performed the in vivo experiments; Wei TANG was in charge of the data analysis; Fa-jun NAN and Jian-ping ZUO designed and supervised the project and wrote the manuscript.
Abbreviations
3TC, lamivudine; ADV, adefovir; CTD, carboxy-terminal domain; DMSO, dimethyl sulfoxide; ETV, entecavir; HBcAg, HBV core protein; HBV, hepatitis B virus; HBVPol, HBV polymerase protein; HPLC, high-performance liquid chromatography; pgRNA, pregenomic RNA.
References
Grimm D, Thimme R, Blum HE . HBV life cycle and novel drug targets. Hepatol Int 2011; 5: 644–53.
Negro F . Management of chronic hepatitis B: an update. Swiss Med Wkly 2011; 141: w13264.
Leemans WF, Ter Borg MJ, de Man RA . Review article: success and failure of nucleoside and nucleotide analogues in chronic hepatitis B. Aliment Pharmacol Ther 2007; 26 Suppl 2: 171–82.
Lucifora J, Zoulim F . The life cycle of hepatitis B virus and antiviral targets. Future Virol 2011; 6: 599–614.
Choi IG, Yu YG . Interaction and assembly of HBV structural proteins: novel target sites of anti-HBV agents. Infect Disord Drug Targets 2007; 7: 251–6.
Chen HJ, Wang WL, Wang GF, Shi LP, Gu M, Ren YD, et al. Rational design and synthesis of 2,2-bisheterocycle tandem derivatives as non-nucleoside hepatitis B virus inhibitors. Chem Med Chem 2008; 3: 1316–21.
Deres K, Schroder CH, Paessens A, Goldmann S, Hacker HJ, Weber O, et al. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 2003; 299: 893–6.
Ying C, Li Y, Leung CH, Robek MD, Cheng YC . Unique antiviral mechanism discovered in anti-hepatitis B virus research with a natural product analogue. Proc Natl Acad Sci U S A 2007; 104: 8526–31.
Wu C, Deng W, Deng L, Cao L, Qin B, Li S, et al. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J Virol 2012; 86: 4658–69.
Newman M, Chua PK, Tang FM, Su PY, Shih C . Testing an electrostatic interaction hypothesis of hepatitis B virus capsid stability by using an in vitro capsid disassembly/reassembly system. J Virol 2009; 83: 10616–26.
Porterfield JZ, Dhason MS, Loeb DD, Nassal M, Stray SJ, Zlotnick A . Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J Virol 2010; 84: 7174–84.
Sells MA, Chen ML, Acs G . Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 1987; 84: 1005–9.
Tong XK, Qiu H, Zhang X, Shi LP, Wang GF, Ji FH, et al. WSS45, a sulfated alpha-D-glucan, strongly interferes with Dengue 2 virus infection in vitro. Acta Pharmacol Sin 2010; 31: 585–92.
Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, et al. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology 2011; 53: 1476–85.
Guan SH, Lu M, Grunewald P, Roggendorf M, Gerken G, Schlaak JF . Interferon-alpha response in chronic hepatitis B-transfected HepG2.2.15 cells is partially restored by lamivudine treatment. World J Gastroenterol 2007; 13: 228–35.
Nguyen DH, Hu J . Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J Virol 2008; 82: 6852–61.
Chua PK, Wang RY, Lin MH, Masuda T, Suk FM, Shih C . Reduced secretion of virions and hepatitis B virus (HBV) surface antigen of a naturally occurring HBV variant correlates with the accumulation of the small S envelope protein in the endoplasmic reticulum and Golgi apparatus. J Virol 2005; 79: 13483–96.
Hu Q, Fang Y, Zhang ZM, Zhang XY, Zhang ZH, Yang DL . Establishment of an in vivo model for duck hepatitis B virus infection using Hubei duckling. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 2008; 22: 113–5.
Schädler S, Hildt E . HBV life cycle: entry and morphogenesis. Viruses 2009; 1: 185–209.
Bourne CR, Finn MG, Zlotnick A . Global structural changes in hepatitis B virus capsids induced by the assembly effector HAP1. J Virol 2006; 80: 11055–61.
Bourne C, Lee S, Venkataiah B, Lee A, Korba B, Finn MG, et al. Small-molecule effectors of hepatitis B virus capsid assembly give insight into virus life cycle. J Virol 2008; 82: 10262–70.
Feld JJ, Colledge D, Sozzi V, Edwards R, Littlejohn M, Locarnini SA . The phenylpropenamide derivative AT130 blocks HBV replication at the level of viral RNA packaging. Antiviral Res 2007; 76 168–77.
Katen SP, Chirapu SR, Finn MG, Zlotnick A . Trapping of hepatitis B virus capsid assembly intermediates by phenylpropenamide assembly accelerators. ACS Chem Biol 2010; 5: 1125–36.
Acknowledgements
We thank Dr Meng-ji LU (Institute of Virology, University Hospital of Essen, Essen, Germany) for data analysis and reading of the manuscript.
The work was supported by the Chinese Academy of Sciences (CAS) Knowledge Innovation Project KSCX1-YW-10-03, National 863 Program Fund 2008AA02Z431, National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” 2009ZX09102-024 and 2012ZX09101-113.
Author information
Authors and Affiliations
Corresponding authors
PowerPoint slides
Rights and permissions
About this article
Cite this article
Yang, L., Shi, Lp., Chen, Hj. et al. Isothiafludine, a novel non-nucleoside compound, inhibits hepatitis B virus replication through blocking pregenomic RNA encapsidation. Acta Pharmacol Sin 35, 410–418 (2014). https://doi.org/10.1038/aps.2013.175
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/aps.2013.175
Keywords
This article is cited by
-
The SAR-based development of small molecular HBV capsid assembly modulators
Medicinal Chemistry Research (2022)
-
Role of Core/Capsid Inhibitors in Functional Cure Strategies for Chronic Hepatitis B
Current Hepatology Reports (2020)
-
Current landscape and future prospects of antiviral drugs derived from microbial products
The Journal of Antibiotics (2018)
-
Drugs in Development for Hepatitis B
Drugs (2017)
-
Prevention of hepatitis B virus infection: from the past to the future
European Journal of Clinical Microbiology & Infectious Diseases (2015)