Abstract
Recent studies show a susceptibility locus (DFNB1) responsible for non-syn-dromic neurosensory autosomal-recessive deafness (NSRD) mapping to the pericentromeric region of chromosome 13q. In order to better understand the frequency with which DFNB1 is the gene for deafness in our patient population and the role of DFNB1 in Caucasians, we performed a genetic linkage study with four microsatellite markers linked to DFNB1 in a total of 48 independent Mediterranean families, of which 30 and 18 were of Italian and Spanish descent, respectively. A maximum two-point lod score of 7.28 was found with marker D13S115 at a recombination frequency of 6 0.1. Significant lod scores were also obtained for D13S143, D13S292 and D13S175. Genetic heterogeneity was confirmed using the HOMOG program which indicated absence of linkage to DFNB1 in approximately 21% of the sample. This study clearly demonstrates that DFNB1 plays an important role in 79% of Mediterranean families with NSRD. Furthermore, results from multipoint analysis predict that the DFNB1 gene maps between markers D13S175 and D13S115 which are separated by approximately 14.2 cM.
Similar content being viewed by others
Introduction
Hereditary hearing loss comprises a broad spectrum of forms ranging from simple deafness to genetically determined syndromes. Non-syndromic neurosensory autosomal-recessive deafness (NSRD) is the most common form of genetic hearing loss accounting for about half of childhood prelingual deafness [1–3]. Over the past few years several studies of large pedigrees of geographically and/or culturally isolated populations have contributed to the mapping of 12 human candidate genes (DFNB) responsible for NSRD. However, limited clinical differentiation and marked genetic heterogeneity make gene(s) identification a challenging task. DFNB1 and DFNB2 were identified by homozygosity mapping of deafness in consanguineous families from Tunisia to chromosomes 13q12 and 11q13.5, respectively [4, 5]. DFNB3 on 17p11.2-q12 was also linked to deafness in consanguineous families from a remote village in Bali [6]. A fourth deafness locus, DFNB4, described in Middle Eastern Druze individuals, was positioned at 7q31 [7]. Three additional candidate genes, DFNB5, DFNB6 and DFNB7, mapped to 14q12, 3p14–p21 and 9q13–q21, respectively, by studying multiple inbred families from India [8–10]. DFNB8 was mapped to the distal arm of chromosome 21 in a family with NSRD from Pakistan [11]; DFNB9 located at 2p22–23, was described in a consanguineous family living in an isolated region of Lebanon [12]. Finally, DFNB10 was recently located in a 12 cM region near the telomere of chromosome 21 in a large inbred Palestinian family [13] and DFNB12 to 10q21–22 in a consanguineous Sunni family from Syria [14].
The DFNB1 locus was recently studied in 19 families of Celtic ancestry, and those results suggest an important contribution of this locus to deafness in the Caucasian population [15]. In contrast, a study in 15 consanguineous NSRD families from Pakistan showed linkage to chromosome 13 in only one kindred [16]. In order to further investigate the role of DFNB1 in NSRD families of Mediterranean descent, we have conducted linkage analysis in 48 families of Mediterranean descent with four markers near the DFNB1 locus. We present here evidence for an important contribution of the DFNB1 locus to hearing impairment in these populations.
Materials and Methods
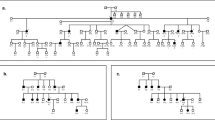
Thirty Italian and 18 Spanish families with deafness were included in the study. They were recruited in several clinical genetics services and schools for the deaf from Southern Italy and Spain. Diagnosis of non-syndromic genetic neurosensory deafness was established according to accepted clinical criteria [17]. Only families with at least 2 affected members and a recessive pattern of inheritance were included in the study. The majority of our families included large pedigrees having several healthy and affected individuals. Only a small proportion was characterized as nuclear families with 2 affected sibs (5 of 30 Italian and 6 of 18 Spanish families). In these cases, although formally possible, dominant inheritance seems unlikely given the absence of deafness elsewhere in the pedigree. We therefore assumed recessive inheritance. Five Italian and 5 Spanish consanguineous families were also included. Examples of family pedigrees included in the study are reported in figure 1. In all cases, care was taken to eliminate families in which pharmaceutical treatment, infection or oto-trauma might have been responsible for deafness. Audiometric evaluations were done on affected individuals and their relatives. Hearing loss was severe or profound at the middle and higher frequencies with a few individuals showing some sparing at lower frequencies. Pure-tone audiometry with air and bone conduction at 125, 250, 500, 1,000, 2,000, 4,000 and 8,000 Hz was performed with a Beltone 2000 audiometer. The younger children were tested for auditory brainstem responses (ABR) using a Lindar apparatus. Early onset of the disease was detected in the first year of life. Differential diagnosis of other forms of non-syndromic deafness, such as Usher syndrome, was considered in all cases but excluded on the basis of clinical, audiometric and laboratory findings.
The total number of individuals available for linkage analysis was 382. The 30 Italian and 18 Spanish families included 76 and 45 affected individuals, respectively, leaving 261 unaffecteds for study. Informed consent was obtained from all subjects and from parents of under-aged patients.
Four chromosome 13 microsatellite markers linked to the DFNB1 locus were investigated in each family using a 373A DNA sequencer and GeneScan software (PE-ABD, Foster City, Calif., USA). High-molecular-weight DNA was extracted from peripheral blood leukocytes using previously described protocols [18] or with a nucleic acid extractor (PE-ABD). PCR primers were synthesized on a 394 DNA Synthesizer (PE-ABD) [19] with fluorochrome-labeled primers prepared by chemically attaching the fluorescent amidite reagent 6-FAM. Deprotection and purification was done using the recommended protocol for dye-labeled oligonucleotides [20]. Primer sequences for D13S175, D13S143, D13S115 and D13S292 were as previously described [21–23]. DNA concentration was adjusted to 40 ng/ml and PCR amplification was performed on an automated thermal cycler (Perkin Elmer, Norvalk, Conn., USA) using conditions previously described for each microsatellite marker. Following PCR amplification, samples were multiplexed at the gel loading step. Electrophoresis was done on 4.75% (w/v) Polyacrylamide gels in 8 M urea containing 1X TBE. Data was collected using GeneScan Data Collection program version 1.1 and analyzed using GeneScan Data Analysis software version 1.2.1b1 or Genotyper version 1.1 (PE-ABD).
Linkage was assessed by analysis of allele segregation of the four different polymorphic markers (D13S175, D13S143, D13S115 and D13S292). Pairwise linkage analysis was performed using MLINK and ILINK from the FASTLINK program package (version 2.2) [24]. Gene frequency was based on NSRD disease prevalence (q = 0.001), and full penetrance was also assumed. Calculations were performed at recombination fractions (Θ) of 0, 0.05, 0.10, 0.20, 0.30 and 0.40 between each marker and the disease locus. Markers allele frequencies were as previously reported (Genome Data Database: https://doi.org/gdbwww.gdb.org/gdb/gdbtop.html).
Multipoint linkage analysis using Linkage LINKMAP software package [25] was performed with the DFNB1 locus against a fixed genetic map with markers D13S175, D13S143, D13S115 and D13S292. Genetic distance between markers was calculated from data obtained from the chromosome 13 map matched with that obtained in our families. Multipoint analyses were done by recoding the markers to five alleles in each family separately. Recombination frequencies were transformed to map distances by Kosambi’s formula and multipoint analyses assumed no interference or sex differences in recombination frequencies [26].
The HOMOG program was used to test for nonallelic heterogeneity using multipoint lod scores calculated for 122 locations of the disease gene at 1-cM intervals within and on either side of the map of markers [26].
Results
To ascertain the frequency with which DFNB1, located on chromosome 13, is the gene responsible for deafness in our patient population, we performed linkage analysis with four microsatellite markers which map close to DFNB1 in 48 large NSRD pedigrees. Pairwise linkage analysis in the 30 Italian families gave positive lod scores with all four markers of chromosome 13 (table 1). The highest linkage was for D13S115 with a lod score of 5.39 at a recombination frequency (Θ) of 0.09. In addition, positive results were obtained with D13S143 (Z = 5.25 at Θ = 0.052). The same analysis performed on the 18 Spanish families indicated D13S175 as the most tightly linked marker (Z = 3.40 at Θ = 0.06) (table 1B). Positive but not significant lod scores were also obtained with the remaining three markers, particularly with D13S115 (Z = 2.02 at Θ = 0.13). Pooling the two populations under study, a lod score of 7.28 at Θ = 0.107 was detected between DFNB1 and D13S115. Significant lod scores were also obtained with the other three markers. These findings clearly demonstrate that DFNB1 plays a major role in determining deafness in our populations.
To define the position of DFNB1 relative to the four markers under study a multipoint linkage analysis was done. The maximum map-specific lod score obtained in multipoint analysis was 8.91, and was found in the interval between D13S175 and D13S115. This location is further supported by a recombinational event detected in two affected members in two unrelated families (data not shown). In both cases the recombinations are in agreement with a DFNB1 gene location centromeric to marker D13S115.
For each of the 122 map positions of the disease locus that were evaluated, a maximum multipoint lod score was calculated under non-allelic heterogeneity by varying alpha, the proportion of linked families (fig. 2). Again, lod scores peaked in the interval between D13S175 and D13S115 (table 2), with a best estimate of alpha of 0.79. From the maximum heterogeneity lod score of 10.54 and the maximum lod score under homogeneity (8.91), odds in favor of heterogeneity were calculated to be 42.7 to 1.A one lod-unit down region was constructed jointly for alpha and the map location, and included alpha values between 0.52 and 0.917, and locations from 3 through 11 cM from D13S175 in the direction of D13S115. There was little difference between the Italian and the Spanish data set with respect to the estimated contribution of the DFNB1 locus: 71 and 77% of families showed linkage, respectively (table 2), when evaluated at the best location for each separate data set, and 80 and 78%, respectively, when evaluated at the best location for the combined data set. Data were analyzed further with the HOMOG2 program, which allows for the presence of two different disease loci within the same map. This analysis yielded only slightly higher lod scores than the previous analyses (Zmax = 10.89), and therefore there appears to be little support for the presence of two deafness loci in the 13q12 region.
Multipoint linkage map assuming a fixed genetic map consisting of markers D13S175, D13S143, D13S115 and D13S292 with respect to locus trait DFNB1, in Italian and Spanish families affected with NSRD. We placed the 0 cM genetic distance value at D13S175. The conversion from recombination fraction values to genetic distances in centi-Morgans has been done according to the Kosambi map function. The peak of the lod score curve favors a location of DFNB1 between D13S175 and D13S115, which encompasses a genetic region of 14.2 cM.
Discussion
This work represents the first large-scale study on linkage analysis to DFNB1 of Mediterranean families with NSRD. DFNB1 was originally mapped in 2 large consanguineous Tunisian kindreds [4]. Its contribution to NSRD in a Pakistani population, from a study of 27 families in the Mirpur region [16], was apparently small, with only 1 family showing linkage. However, a study based on 19 Anglo-Saxon/Celtic families from New Zealand and Australia indicated that a more substantial fraction of NSRD in this population might be due to a mutation at the DFNB1 locus [15]. While statistically significant linkage was not demonstrated for D13S175, D13S143 and D13S115 in this study, nine families showed cosegrega-tion of marker haplotypes with deafness, indicating that the DFNB1 locus contributes to a fraction of congenital NSRD in individuals of Anglo-Saxon/Celtic ancestry. In this report we clearly demonstrate that DFNB1 plays an important role in determining deafness in Italian and Spanish patients. In both the Italian and the Spanish NSRD data sets, a large and strikingly similar proportion of families (80 and 78%, respectively) show linkage to DFNB1. The observed odds in favor of nonallelic heterogeneity (42.7 to 1) show that other genes must be involved in determining NSRD in these populations. It is not clear if a single additional gene or multiple genes account for the remaining 21% of the population. Our preliminary data on DFNB2 and DFNB4 suggest that DFNB4 accounts for some of the additional cases of NSRD.
With respect to the location of DFNB1, our results suggest that its most likely position maps between D13S175 and D13S115, which were found to be 14.2 cM apart. This location is further supported by two recombinational events detected in affected members from two unrelated families. In both cases the recombinations are in agreement with a DFNB1 gene location centromeric to the marker D13S115.
Work is now focused on analysis of additional chromosome 13 markers in DFNB1-linked families to further narrow and refine the location of the DFNB1 gene. Definition of the two recombinants and identification of additional ones will help in defining the exact location of the DFNB1 gene. In addition, work continues to determine how many and which loci are linked to NSRD in those families which do not show linkage to DFNB1.
References
McKusick VA: Mendelian Inheritance in Man, ed. 10. Baltimore, Johns Hopkins University Press, 1992.
Reardon W: Genetic deafness. J Med Genet 1992;29:521–526.
Steel HP, Brown SDM: Genes and deafness. Trends Genet 1994;10:428–435.
Guilford P, Arab SB, Blanchard S, Levilliers J, Weissenbach J, Belkahia A, Petit C: A non-syndromic form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nature Genet 1994;6:24–28.
Guilford P, Ayadi H, Blanchard S, Chaib H, Le Paslier D, Weissenbach J, Drira M, Petit C: A human gene responsible for neurosensory, non-syndromic recessive deafness is a candidate homologue of the mouse sh-1 gene. Hum Mol Genet 1994;3:989–993.
Friedman TB, Liang Y, Weber JL, Hinnant JT, Barber TD, Winata S, Arhya IN, Asher JH Jr: A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nature Genet 1995;9:86–91.
Baldwin CT, Farrer LA, Weiss S, De Stefano AL, Adair R, Franklyn B, Kidd KK, Korostishevsky M, Bonne-Tamir B: Linkage of congenital, recessive deafness (DFNB4) to chromosome 7q31 and evidence for genetic heterogeneity in the Middle Eastern Druze population. Hum Mol Genet 1995;4:1637–1642.
Fukushima K, Arabandi R, Srisailapathy CRS, Ni L, Chen A, O’Neill M, Van Camp G, Coucke P, Smith SD, Kenyon JB, Jain P, Wilcox ER, Zbar RIS, Smith RJH: Consanguineous nuclear families used to identify a new locus for recessive non-syndromic hearing loss on 14q. Hum Mol Genet 1995;4:1643–1648.
Fukushima K, Arabandi R, Srisailapathy CRS, Ni L, Wayne S, O’Neill ME, Van Camp G, Coucke P, Jain P, Wilcox ER, Smith SD, Kenyon JB, Zbar RIS, Smith RJH: An autosomal recessive non-syndromic form of sensorineural hearing loss maps to 3p-DFNB6. Genome Res 1995;5:305–308.
Jain PK, Fukushima K, Deshmukh D, Arabandi R, Thomas E, Kumar S, Lalwani AK, Kumar S, Ploplis B, Skarka H, Srisailapathy CRS, Wayne S, Zbar RIS, Verma IC, Smith RJH, Wilcox ER: A human recessive neurosensory nonsyndromic hearing impairment locus is a potential homologue of the murine deafness (dn) locus. Hum Mol Genet 1995;4:2391–2394.
Veske A, Oehlmann R, Younus F, Mohyuddin A, Muller-Myhsok B, Qasim Mehdi S, Gal A: Autosomal recessive non-syndromic deafness locus (DFNB8) maps on chromosome 21q22 in large consanguineous kindred from Pakistan. Hum Mol Genet 1996;5:165–168.
Chaib H, Place C, Salem N, Chardenoux S, Vincent C, Weissenbach J, El-Zir E, Loiselet J, Petit C: A gene responsible for a sensorineural nonsyndromic recessive deafness maps to chromosome 2p22–23. Hum Mol Genet 1996;5: 155–158.
Bonne’-Tamir B, DeStefano AL, Briggs CE, Adair R, Franklyn B, Weiss S, Korostishevsky M, Frydman M, Baldwin CT, Farrer LA: Linkage of congenital recessive deafness (gene DFNB10) to chromosome 21q22.3. Am J Hum Genet 1996;58:1254–1259.
Chaib H, Place C, Salem N, Salem N, Dode’ C, Chardenoux S, Weissenbach J, El-Zir E, Loiselet J, Petit C: Mapping of DFNB12, a gene for a non-syndromal autosomal recessive deafness, to chromosome 10q21–22. Hum Mol Genet 1996;5:1061–1064.
Maw MA, Allen Powell DR, Goodey RJ, Stewart IA, Nancarrow DJ, Hayward N, McKinlay Gardner RJ: The contribution of the DFNB1 locus to neurosensory deafness in a Caucasian population. Am J Hum Genet 1995; 57:629–635.
Brown KA, Janjua AH, Karbani G, Parry G, Noble A, Crockford G, Bishop DT, Newton VE, Markham AF, Mueller RF: Linkage studies of non-syndromic recessive deafness (NSRD) in a family originating from the Mirpur region of Pakistan maps DFNB1 centromeric to D13S175. Hum Mol Genet 1996;5: 169–173.
Nance WE, Sweeney A: Genetic factors in deafness of early life. Otolaryngol Clin North Am 1975;8:19–48.
Poncz M, Solowiejzcyk D, Harpel B, Mory Y, Schwartz E, Surrey S: Construction of human gene libraries from small amounts of peripheral blood: Analyses of β-like globin genes. Hemoglobin 1982;6:27–36.
Caruthers MH, Barone AD, Beaucage SL, Dodds DR, Fisher EF, McBride LJ, Matteucci M, Stabinsky Z, Tang J-Y: Chemical synthesis of deoxyoligonucleotides by the phosphoram-idite method. Methods Enzymol 1987; 154: 287–313.
Applied Biosystems, Inc: Synthesis of fluorescent dye-labeled oligonucleotides for use as primers in fluorescent-based DNA sequencing. ABI User Bull 1991;11:6–9.
Hudson TJ, Engelstein M, Lee MK, Ho EC, Rubenfield MJ, Adams CP, Housman DE, Dracopoli NC: Isolation and chromosomal assignment of 100 highly informative human simple sequence repeat polymorphisms. Genomics 1992;13:622–629.
Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseiz G, Lathrop M: A second-generation linkage map of the human genome. Nature 1992;359:794–801.
Petrukhin KE, Speer MC, Cayanis E, de Fatima Bonaldo M, Tantravahi U, Soares MB, Fischer SG, Warburton D, Gilliam TC, Ott J: A microsatellite genetic linkage map of human chromosome 13. Genomics 1993;15:76–85.
Cottingam R, Idury R, Schaffer A: Faster sequential genetic linkage computations. Am J Hum Genet 1993;53:252–263.
Lathrop GM, Lalouel JM, Julier C, Ott J: Multilocus linkage analysis in humans: Detection of linkage and estimation of recombination. Am J Hum Genet 1984;37:482–498.
Ott J: Analysis of Human Genetic Linkage. Baltimore, Johns Hopkins University Press, 1991.
Acknowledgments
We are indebted to families for their cooperation and the Studio Logopedico Elvira Signori. The authors thank Dr. Eric Rappaport for his helpful advice in microsatellite analyses. This work was supported in part by a grant from Italian Ministry of Health (P.G.), the Catalan Heath Service (X.E. and V.V.), a ‘Fundación Ramón Areces’ research grant (X.E., N.G. and S.C.) and by the Department of Pathology, The Children’s Hospital of Philadelphia (P.F.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gasparini, P., Estivill, X., Volpini, V. et al. Linkage of DFNB1 to Non-Syndromic Neurosensory Autosomal-Recessive Deafness in Mediterranean Families. Eur J Hum Genet 5, 83–88 (1997). https://doi.org/10.1007/BF03405882
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF03405882