Abstract
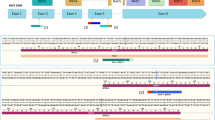
Previous studies have suggested that differences in the ability of normal and malignant cells to process certain alternatively spliced pre-mRNA transcripts can be exploited as a potentially powerful means of targeting the expression of therapeutic genes to tumor cells in vivo and in vitro. Specifically, it was shown that efficient processing of minigene constructs containing the alternatively spliced CD44 exons v9 and v10 only occurs in tumor cells that express CD44 isoforms that incorporate these exons (e.g. CD44R1). In the present study, efforts were made to define the molecular mechanisms that underlie the apparent specificity of this process. RT-PCR analysis and DNA sequencing were used to characterize the various splicing events that occur between CD44 exons v8, v9 and v10 following transfection of minigene constructs containing these various exons into CD44R1-positive (PC3) and CD44R1-negative (T24) cell lines. The results obtained confirm that although the v8–v9 intron is efficiently removed in both CD44R1-positive and CD44R1-negative cells, the corresponding v9–v10 intron is accurately spliced and the exons appropriately joined only in lines that express v10-containing CD44 isoforms (e.g. PC3). In CD44R1-negative cell lines (e.g. T24) alternative 5′ and 3′ splice sites located within the v9–v10 intron are preferentially used, resulting in various portions of the intron being retained within the final processed mRNA product. It is proposed that identification of these functionally important intronic sequence elements will facilitate the development of second generation “splice activated gene expression” vectors that may prove useful in various cancer gene therapy applications.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Green NK, Seymour LW . Adenoviral vectors: systemic delivery and tumor targeting. Cancer Gene Ther. 2002;9:1036–1042.
Greco O, Marples B, Dachs GU, et al. Novel chimeric gene promoters responsive to hypoxia and ionizing radiation. Gene Therapy. 2002;9:1403–1411.
Greco O, Scott SD, Marples B, et al. Cancer gene therapy: ‘delivery, delivery, delivery’. Front Biosci. 2002;7:d1516–d1524.
Hayes GM, Carpenito C, Davis PD, et al. Alternative splicing as a novel of means of regulating the expression of therapeutic genes. Cancer Gene Ther. 2002;9:133–141.
Dreyfuss G, Hentze M, Lamond AI . From transcript to protein. Cell. 1996;85:963–972.
Lopez AJ . Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305.
Cooper DL, Dougherty GJ . To metastasize or not? Selection of CD44 splice sites. Nat Med. 1995;1:635–637.
Naot D, Sionov RV, Ish-Shalom D . CD44: structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319.
Dougherty GJ, Landorp PM, Cooper DL, et al. Molecular cloning of CD44R1 and CD44R2, two novel isoforms of the human CD44 lymphocyte “homing” receptor expressed by hemopoietic cells. J Exp Med. 1991;174:1–5.
Sneath RJ, Mangham DC . The normal structure and function of CD44 and its role in neoplasia. Mol Pathol. 1998;51:191–200.
Chiu RK, Droll A, Dougherty ST, et al. Alternatively spliced CD44 isoforms containing exon v10 promote cellular adhesion through the recognition of chondroitin sulfate-modified CD44. Exp Cell Res. 1999;248:314–321.
Kaighn ME, Lechner JF, Narayan KS, et al. Prostate carcinoma: tissue culture cell lines. Natl Cancer Inst Monogr. 1978;49:17–21.
Kaighn ME, Narayan KS, Ohnuki Y, et al. Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Invest Urol. 1979;17:16–23.
O'Toole C, Perlmann P, Unsgaard B, et al. Cellular immunity to human urinary bladder carcinoma. I. Correlation to clinical stage and radiotherapy. Int J Cancer. 1972;10:77–91.
Dougherty GJ, Cooper DL, Memory JF, et al. Ligand binding specificity of alternatively spliced CD44 isoforms. Recognition and binding of hyaluronan by CD44R1. J Biol Chem. 1994;269:9074–9078.
Fu XD . The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680.
Adams MD, Rudner DZ, Rio DC . Biochemistry and regulation of pre-mRNA splicing. Curr Opin Cell Biol. 1996;8:331–339.
Mayeda A, Krainer AR . Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375.
Mayeda A, Munroe SH, Caceres JF, et al. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 1994;13:5483–5495.
Smith CW, Valcarcel J . Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25:381–388.
Screaton GR, Caceres JF, Mayeda A, et al. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349.
Galiana-Arnoux D, Lejeune F, Gesnel MC, et al. The CD44 alternative v9 exon contains a splicing enhancer responsive to the SR proteins 9G8, ASF/SF2, and SRp20. J Biol Chem. 2003;278:32943–32953.
Konig H, Ponta H, Herrlich P . Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913.
Bell MV, Cowper AE, Lefranc MP, et al. Influence of intron length on alternative splicing of CD44. Mol Cell Biol. 1998;18:5930–5941.
Yu Q, Toole BP . A new alternatively spliced exon between v9 and v10 provides a molecular basis for synthesis of soluble CD44. J Biol Chem. 1996;271:20603–20607.
Mironov AA, Fickett JW, Gelfand MS . Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293.
Hanke J, Brett D, Zastrow I, et al. Alternative splicing of human genes: more the rule than the exception? Trends Genet. 1999;15:389–390.
Caballero OL, de SSJ, Brentani RR, et al. Alternative spliced transcripts as cancer markers. Dis Markers. 2001;17:67–75.
Chiu RK, Carpenito C, Dougherty ST, et al. Identification and characterization of CD44RC, a novel alternatively spliced soluble CD44 isoform that can potentiate the hyaluronan binding activity of cell surface CD44. Neoplasia. 1999;1:446–452.
Tasch J, Gong M, Sadelain M, et al. A unique folate hydrolase, prostate-specific membrane antigen (PSMA): a target for immunotherapy? Crit Rev Immunol. 2001;21:249–261.
Kong QY, Liu J, Chen XY, et al. Differential expression patterns of hyaluronan receptors CD44 and RHAMM in transitional cell carcinomas of urinary bladder. Oncol Rep. 2003;10:51–55.
Toran-Allerand CD . Mini-review: a plethora of estrogen receptors in the brain: where will it end? Endocrinology. 2004;145:1069–1074.
Leung DW, Tompkins CK, White T . Characterization of two spliced variants of human phosphatidic acid phosphatase cDNAs that are differentially expressed in normal and tumor cells. Adv Exp Med Biol. 1999;469:639–646.
Nichols SE, Harper DC, Berson JF, et al. A novel splice variant of Pmel17 expressed by human melanocytes and melanoma cells lacking some of the internal repeats. J Invest Dermatol. 2003;121:821–830.
Lee JH, Seo YW, Park SR, et al. Expression of a splice variant of KAI1, a tumor metastasis suppressor gene, influences tumor invasion and progression. Cancer Res. 2003;63:7247–7255.
Lu F, Gladden AB, Diehl JA . An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003;63:7056–7061.
Philips AV, Cooper TA . RNA processing and human disease. Cell Mol Life Sci. 2000;57:235–249.
Sampath J, Long PR, Shepard RL, et al. Human SPF45, a splicing factor, has limited expression in normal tissues, is overexpressed in many tumors, and can confer a multidrug-resistant phenotype to cells. Am J Pathol. 2003;163:1781–1790.
Acknowledgements
This work was supported, in part, by Grant RO1 CA100004 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hayes, G., Dougherty, S., Davis, P. et al. Molecular mechanisms regulating the tumor-targeting potential of splice-activated gene expression. Cancer Gene Ther 11, 797–807 (2004). https://doi.org/10.1038/sj.cgt.7700759
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cgt.7700759
Keywords
This article is cited by
-
Exploiting the tumor microenvironment in the development of targeted cancer gene therapy
Cancer Gene Therapy (2009)
-
Loss of Endocan tumorigenic properties after alternative splicing of exon 2
BMC Cancer (2008)
-
Identification of a human TFPI-2 splice variant that is upregulated in human tumor tissues
Molecular Cancer (2007)