Abstract
A population analysis of an unusual NOR-site polymorphism previously detected in brown trout (Salmo trutta L.) from North-western Spain was carried out in 225 individuals from 20 native populations from this area. The analysis performed has permitted us to reveal: (i) the ubiquity of this phenomenon in most river basins from NW Spain, 13 extra-NORs not observed in standard trout being detected; (ii) the rDNA constitution of all extra-NORs, as confirmed by CMA3-staining and rDNA-FISH, and their capability to constitute their own nucleolus; (iii) the constant location of extra-NORs within individuals and their stable transmission across generations; and (iv) the telomeric location of Ag-NORs, which were randomly distributed in the karyotype of S. trutta, mostly in the heterozygous condition. Repetitive sequences in the IGS or scattered along rDNA units, rather than chromosome rearrangements, could play an important role in the dispersion of NORs. The polymorphism described seems to have a single geographical origin, since a positive correlation was demonstrated between the degree of polymorphism and geographical distance to a central point of the river basin where the phenomenon showed a higher intensity. The distribution of NOR-site variation as compared with allozyme variation in the populations studied, showed significant statistical differences. Selection against high copy number, or mutation due to changes in NOR location, could be acting on this polymorphism to explain the observed distribution. The high dispersion of NOR-site variants within and among populations suggests the transposition phenomenon responsible for NOR jumping could be still active.
Similar content being viewed by others
Introduction
In eukaryotes the genes coding for the 18S-5.8S-28S rRNA are organized in long tandem arrays separated by intergenic spacers (IGS), which contain the regulatory elements for rDNA transcription (Moss & Stefanovsky, 1995), and are clustered at specific chromosome sites called nucleolar organizer regions (NORs). NORs are very dynamic regions in evolutionary terms, both in a macro as well as in a microevolutionary view. Consequently, these regions have been frequently used as phylogenetic markers (Amemiya & Gold, 1988), differences in chromosome location being detected even between sibling species (Volleth, 1987). These changes in position during evolution have been quite often attributed to chromosome rearrangements (Hall & Parker, 1995).
On the other hand, conventional cytogenetic and the most recent hybridization techniques have shown NOR regions to be also polymorphic both in number and location within species (Schmid et al., 1995; Zhuo et al., 1995). Although one NOR-bearing chromosome pair is usually considered plesiomorphic in most groups analysed, some vertebrate species show a multichromosomal location of NORs (Suzuki et al., 1990; Schmid et al., 1995). A constant number of several stable NOR sites has been usually observed in these species, but in some cases, the multichromosomal pattern appears to be unstable.
A common multichromosomal pattern described in amphibian (De Lucchini et al., 1993), fish (Zhuo et al., 1995) and plant (Schubert & Wobus, 1985) species involves a main NOR-bearing chromosome pair present in all individuals, and other highly variable sites, usually in heterozygous condition. In these examples, NOR regions seem capable of spreading throughout the genome creating new rDNA loci. Different mechanisms, such as transposable elements adjacent to ribosomal genes (Schubert & Wobus, 1985), repetitive elements which could serve as points for chromosome exchange (Maggini et al., 1991), amplification of minor rDNA loci (Dubcovsky & Dvörak, 1995), and reinsertion of extrachromosomal rDNA amplified during oogenesis (Phillips et al., 1988), have been proposed to explain the apparent mobility of NORs. To which extent these unstable multichromosomal patterns could reflect transitional evolutionary events, heading towards a new stable location of NORs, remains as an open question.
Studies of chromosome location of NORs in salmonids have shown a single NOR-bearing chromosome pair in most species analysed (Phillips & Ihssen, 1985; Phillips et al., 1986). Only species of the genus Salvelinus have shown a large multichromosomal NOR distribution, usually unstable (Phillips et al., 1988; Zhuo et al., 1995). The situation reported in S. trutta represents an intermediate position between the multichromosomal location of NORs in Salvelinus and the presence of a single NOR-bearing chromosome pair of most salmonids. Active NORs in brown trout (standard individuals) are located on the short arms of pair 11, and occasionally on a medium subtelocentric pair (Martínez et al., 1991). Minor rDNA clusters have also been observed in several subtelocentric pairs by means of CMA3-staining and rDNA-FISH, but are always inactive (Martínez et al., 1991; Castro et al., 1996).
In a recent study, we reported an atypical NOR-site polymorphism in brown trout from a river basin in North-western Spain. Specimens from this basin showed an unstable multichromosomal NOR distribution largely different from standard individuals (Castro et al., 1994, 1996). At least 10 atypical active NOR sites not described previously were observed in this river basin, some of them constituting new rDNA clusters, as shown by rDNA-FISH (Castro et al., 1996). In contrast with related reports in other species, the polymorphism described in our study was apparently restricted to a single river basin. Brown trout from this area could represent suitable material to deal with basic questions related with the origin and evolutionary dynamics of NOR-site polymorphisms, due to the magnitude of the transposition event detected, its geographical restriction, and possibly, its recent evolutionary origin.
In the present study we characterize in detail the NOR-site distribution in a large number of populations from the Miño and other neighbouring basins in NW Spain. The goal of our study was to advance the knowledge of the mechanism underlying the apparent jumping of NORs in these populations, to obtain some clues about its temporal and spatial origin, and to analyse the evolutionary forces influencing this polymorphism.
Materials and methods
Sampling
Two hundred and twenty-five individuals of S. trutta were collected by electrofishing from 20 native populations from the four main river basins in North-western Spain (Miño, Umia, Tambre and Navia). Three populations previously studied (Cabalar, Tambre and Navia; Castro et al., 1994) were included in the analysis to provide a more reliable picture of NOR-site variation in the area under study (Fig. 1). Between 10 and 15 individuals were analysed in most populations (Table 1). Since the NOR-site polymorphism had been previously detected in the Miño basin (Castro et al., 1994, 1996), the major sampling effort was carried out in this river basin, which is also the largest one in NW Spain (Fig. 1). Some populations from neighbouring river basins were also analysed to assess the extent of this unusual polymorphism. To get some clues about the temporal origin of this polymorphism, three populations isolated by impassable waterfalls were sampled in the Miño basin (Deva, Fondós and Requeixo). A stock of North European origin (Monasterio de Piedra) was analysed as a control, since its NOR distribution conforms to the standard pattern. All populations studied had been previously analysed for 33 allozyme loci (Bouza et al., 1999), which provided a reference to which we could contrast the pattern of NOR-site variation observed. We assumed a neutral hypothesis to deal with allozyme variation in our study, as usually invoked for allozyme markers, but especially in groups like salmonids with small effective population size (Chakraborty & Leimar, 1987; Jorde & Ryman, 1995).
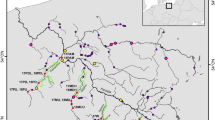
Location of the 20 brown trout natural populations analysed in the present study. Dark spots: non-isolated populations; dark squares: isolated populations; empty symbols: populations analysed in the previous study by et al.(1994). Codes are indicated in Table 1. The arrow points to the junction between the Miño and Sil (its main tributary) rivers, which was considered as a focus for looking for a cline of the NOR-site polymorphism.
Cytogenetic techniques
Ag-staining Chromosome spreads were obtained from kidney culture as described by Martínez et al. (1993). Staining with AgNO3 was performed according to Howell & Black (1980) with slight modifications to reveal the presence of active rDNA clusters. Around 50 plates and 100 nuclei were studied in each individual to obtain the distribution of the number of Ag-NORs per metaphase and the number of nucleoli per nucleus, respectively. In order to establish the Ag-NOR chromosome correspondence among plates within individuals and among individuals and populations, we followed a conservative approach, since only five chromosome pairs (1, 8–9, 10, 11 and 12) can unambiguously be identified in the complex karyotype of S. trutta (2n=80; Martínez et al., 1991). Therefore, Ag-NOR chromosomes different from the five identifiable pairs were classified according to their sizes within each group and numbered from the largest to the smallest. To standardize chromosome measurements among different plates and individuals, the size of each NOR-bearing chromosome was compared to an easily identifiable chromosome structure in the karyotype of S. trutta, namely the long arm of chromosome pair 11. In this way, metacentric chromosomes were designated as M1, M2 and M3, subtelocentric ones as S1, S2 and S3 and acrocentrics as A1, A2 and A3.
Chromomycin A3-staining and rDNA-FISH Chromomycin-staining (CMA3) was performed according to the procedures of Schweizer (1979). Slides from at least five individuals per population were treated with 0.5 mg/mL CMA3 for 2 h and counterstained with 0.5 mg/mL DA for 15 min. In our previous work we could demonstrate the similar resolution of CMA3-staining as compared to rDNA-FISH to reveal rDNA clusters in brown trout (Castro et al., 1996). Thus, we usually applied CMA3-staining, a more straightforward technique, to analyse the structural nature of Ag-NOR sites; i.e. to confirm they are constituted by rDNA clusters. FISH, as described by Castro et al. (1996), was only applied in some cases to obtain a definitive confirmation of rDNA composition of particular Ag-NORs. To establish the chromosome correspondence among individuals and techniques, the same principles outlined before for Ag-NORs were applied.
Statistical analysis
The population analysis of the NOR-site polymorphism in brown trout was carried out mainly starting from Ag-staining data. All Ag-NORs in our study were demonstrated to be constituted by rRNA genes. Therefore the detection of a positive signal at a specific position in more than one plate with Ag-staining was scored as a NOR site in the specimen analysed (Table 2). Also, most rDNA clusters in S. trutta detected with rDNA-FISH and CMA3-staining showed NOR activity at variable frequencies (present work; Castro et al., 1996). In this way, the analysis of a large number of Ag-stained plates per individual permitted us to detect most, if not all, NOR sites in each individual. Therefore, the Ag-NOR pattern was analysed in around 50 metaphases per individual.
We focused our attention on new NOR-sites (extra-NORs) not present in standard individuals, because we were interested in the structural variation rather than in functional phenomena derived from this polymorphism (Castro et al., 1996). Therefore, active NORs in the short arms of several subtelocentric chromosomes, which represented silent positions in standard individuals (Martínez et al., 1991; Castro et al., 1996), were not taken into account in the statistical analysis. Also, when the two homologues of a given extra-NOR bearing chromosome pair were detected in the same individual, they were scored as a single extra-NOR position. The number of the different extra-NOR chromosomes observed in each specimen (nt; Table 2), its range and population mean (nT), the number of extra-NORs per population (T), and the percentage of individuals with extra-NORs, were scored in each population (Table 1). Also, the frequencies of the extra-NORs detected in the populations studied were calculated.
As outlined before, we compared the distribution of NOR-site and allozyme variation within and among populations in the samples analysed in our study. To estimate genetic differentiation among populations, both genetic distances between all population pairs, and the distribution of genetic diversity within and between populations, were calculated. To apply this statistical approach to NOR-site variation, we considered each extra-NOR chromosome pair as a locus. Within each locus, the presence/absence and different location of NORs were counted as its alleles. Starting from these data, we estimated the relative component of genetic differentiation among populations (GST, Nei, 1977; NEGST program, Chakraborty et al., 1982) after excluding isolated samples. The lack of migration to these samples suggested their exclusion to analyse the distribution of genetic diversity in the samples studied. Genetic differentiation between all pairs of populations was estimated by means of Nei’s genetic distance (D; nei, 1978), and the corresponding matrix of distances was constructed. To display genetic relationships among samples a dendrogram was obtained from the D matrix using the Neighbour-Joining (NJ) method (Saitou & Nei, 1987; DISPAN). The same analysis had been previously performed for allozyme data in the same populations (Bouza et al., 1999). Non-parametric tests were performed to compare the values of GST obtained from NOR and allozyme data (Wilcoxon signed-rank test) and to check for a cline observed for NOR-site variation (Spearman correlation). Also, Mantel randomization tests (1000 permutations) were performed to obtain the correlations between genetic and geographical distances to test for isolation by distance, and between the matrices of genetic distances obtained both from NOR and allozyme data, to check for their similarity.
Results
Individual analysis
By combining Ag- and CMA3-staining, and rDNA fluorescent in situ hybridization (FISH) we analysed NOR-site variation in 265 individuals of S. trutta from 23 native populations sampled in North-western Spain (including the three samples previously analysed by Castro et al. (1994)), as well as in 12 specimens of a standard hatchery stock (M. de Piedra). Standard individuals showed the NOR pattern commonly described: conspicuous silver deposits in both members of pair 11, and an occasional third Ag-NOR on a medium subtelocentric pair. The application of CMA3-staining and rDNA-FISH also showed the presence of several silent rDNA clusters in the short arms of several subtelocentric pairs (Martínez et al., 1991).
Polymorphic individuals showed a multichromosomal NOR distribution largely different from standard ones. Up to 20 different NOR positions were observed in polymorphic trout after the application of Ag-staining. A very good correspondence between the frequency distributions of the number of Ag-NORs per metaphase and the number of nucleoli per nucleus was observed in all individuals and populations analysed (Table 3). So, the maximum number of Ag-NOR and nucleoli per cell was the same in most specimens. All Ag-NORs were found at telomeric positions, and showed positive signals after CMA3-staining and rDNA-FISH, which supported their structural nature; that is to say, they appeared to be constituted by rDNA clusters. In most individuals analysed, the extra-NOR chromosomes were observed in heterozygous condition. Many of these sites were not present in standard individuals, so we termed them as extra-NORs, and defined new NOR-bearing chromosomes (11b, M1s, M1l, M1b, M2, M3, 8/9s, 8/9l, 12l, 12s, S1b, A1, A2 and A3; Fig. 2; Table 4). Those specimens with extra-NORs were considered as atypical (Table 1). No variation with regard to the number and position of NORs was observed within individuals after applying CMA3-staining to 107 specimens pertaining to the 20 native populations studied, which indicates the stable location of extra-NORs. Also, several extra-NORs were observed in different individuals from the same population, sometimes at high frequencies (A2; Table 2), which suggests their stable transmission across generations, at least in the short term.
NOR-bearing chromosomes defined by the presence of extra-NORs detected with Ag- (upper row) and CMA3-staining (lower row) in the brown trout populations analysed in this study. Chromosomes are arranged according to centromeric positions. Lowercase letters s, l and b indicate the presence of extra-NORs in the short, long, and both arms of the chromosome, respectively.
Population analysis
Geography of the NOR-site polymorphism The mean (nT) and range of the number of extra-NORs per individual (nt), the total number of extra-NORs per population (T), and the percentage of atypical individuals in each of the 20 native samples and the standard hatchery stock analysed in this study are summarized in Table 1. The number of extra-NORs per individual (nt) ranged from zero to a maximum of four in the most polymorphic specimens. The range of T was from zero in standard samples (Moia, Lamas, M. Piedra) up to nine in the highest polymorphic sample (Bibei; Table 1). Most polymorphic individuals were detected in the Miño basin where atypical fish constituted above 50% of the individuals sampled in all populations studied, excluding Nogueira and Fondós (which only showed standard individuals; Table 1). Also, the mean number of extra-NORs per basin was clearly higher in Miño (T mean=5.09 ± 0.76; excluding the isolated samples) than in Umia (T mean = 2.50 ± 1.50), Tambre (T mean = 1.75 ± 1.18), and Navia (T mean = 0). The highest figures (nT, T, % atypical individuals) were reached in populations located in the central part of the Miño basin, some of these samples (Samos, Bibei, and Sarria) containing only polymorphic individuals and having nT figures above 1.6 (Table 1; Fig. 1). Samples from the ends of the basin, especially the outlet, showed a lower number of extra-NORs per individual (nT) and fewer atypical trout. Isolated samples, together with Nogueira, showed the lowest number of extra-NORs and percentage of polymorphic individuals.
The neighbouring drainages analysed (Umia and Tambre) were also affected by this polymorphism, although to a minor extent. The samples from Umia and Tambre analysed in this study contained polymorphic fish, but in general with lower values of polymorphism. All trout from the three populations sampled in the northernmost river basin (Navia) showed a standard NOR pattern.
These results suggested the existence of a gradient in the degree of the polymorphism, both from the central part to the ends of Miño basin, and from this river basin to the northern ones. To test this hypothesis we carried out a nonparametric Spearman correlation between the population mean of the number of extra-NORs (nT) and the geographical distance to a point located in the central part of the Miño basin (the junction between the Miño river and the Sil river, its main tributary; Fig. 1), where the most highly polymorphic populations were detected. The naturally isolated samples (Fondós, Requeixo and Deva) were excluded from this analysis, because the lack of migration from other samples probably determines lower values of polymorphism. A highly significant negative correlation (rs=−0.64; P=0.002) was observed when applying this test: the more distantly a population was located from this point, the less polymorphic it appeared.
Genetic differentiation within and among populations
When comparing the frequency of extra-NORs within and among populations, a great dispersion of NOR variants was observed. Several extra-NOR chromosomes were detected in a few populations (M1b, 11l, M3, 8–9l, 12s, 12l), whereas only three out of 14 were observed in more than 50% of the atypical samples analysed (M1s, A1, and A2; Fig. 2; Table 4). Also, in most cases, a low frequency of extra-NOR variants was observed within populations. Quite frequently a specific position was evident in only one individual in many samples analysed. For example, 33.44% of extra-NORs in the four most highly polymorphic populations from Miño were detected in one specimen. An example of this observation is presented in Table 2, corresponding to the distribution of extra-NORs in the Ferreira population.
Genetic relationships among populations for NOR-site variation were analysed and compared with those obtained from allozyme data. In order to obtain an estimation of the genetic differentiation among the populations sampled based on NOR variation, the relative component of genetic differentiation GST and Nei’s genetic distances were calculated using NOR data. In this approach we considered each extra-NOR pair as a locus and the presence/absence of Ag deposits and their different positions as its alleles (Table 4).
The genetic differentiation among populations for NOR data (GST=12.4) was very similar and not significantly different from to that obtained from allozyme data (GST=13.6; Wilcoxon signed-rank test; r=−0.178; P=0.859). In spite of the large number and dispersion of extra-NORs, these appeared in low frequency within populations, therefore contributing only a small proportion to the total genetic differentiation among populations.
Neighbour-Joining dendrograms (NJ), computed starting from the matrices of genetic distances between all population pairs both for allozyme and NOR-site variation, were obtained to analyse genetic relationships among populations. As shown in Fig. 3A, when allozymes were used, the highest divergence was observed for isolated populations (Fondós, Deva). The remaining populations appeared clustered according to the geographical distance among them. In fact, a significant correlation was observed between genetic and geographical distances when the isolated samples and the largely divergent population of Nogueira were excluded from the analysis (r=0.332; P=0.018). This picture was sharply different from that obtained for NOR-site variation (Fig. 3B), a nonsignificant correlation being observed between geographical and genetic distances in this case (r=−0.094; P=0.331). The Bibei sample, one of the most polymorphic and not isolated, appeared placed far apart from the rest as a consequence of the high frequency of the M1l extra-NOR chromosome (Fig. 2; Table 4). The remaining samples appeared clustered mainly in accordance to their degree of NOR-site polymorphism. The dissimilarity of genetic distances between allozyme and NOR data was statistically confirmed by analysing the correlation between both matrices, a nonsignificant correlation being obtained after applying a Mantel randomization test (r=−0.137; P=0.220).
Genetic relationships among the populations of brown trout analysed by the Neighbour-Joining (NJ) algorithm (Saitou & Nei, 1987) computed from genetic distances (Nei, 1978) calculated both from (A) allozyme (Bouza et al., 1999) and (B) NOR data.
Discussion
Nature and dispersion mechanism of extra-NORs
In the present work we have carried out an exhaustive population characterization of an unstable multichromosomal NOR-site polymorphism previously described in brown trout from North-western Spain (Castro et al., 1994, 1996). The analysis of 265 individuals from 23 native populations and 12 specimens from a standard hatchery stock, using different cytogenetic techniques, has permitted us to obtain a more accurate characterization of this phenomenon and confirm some hypotheses reported in our previous works. (i) The multichromosomal NOR-site distribution is not an exception but the rule in most of the populations analysed in NW Spain, especially in the Miño basin. We could detect, in addition to the standard positions, up to 13 different extra-NORs (not present in standard individuals), although the conservative approach followed has probably underestimated the actual number of NOR-bearing chromosomes. (ii) All available data strongly indicate that extra-NORs detected in S. trutta in our work correspond to active rDNA clusters. They appear stained with Ag- and CMA3-staining, a good substitute for rDNA-FISH in this species (Castro et al., 1996), and hybridized with probes of rDNA coding regions. It is possible that sequences inserted in rRNA genes, like those described in the most polymorphic population (Castro et al., 1997), could inactivate some of them. However, the very good correspondence between the frequency distributions of the number of Ag-NORs and nucleoli per cell within each individual, clearly suggest that extra-NORs in S. trutta are transcriptionally active and capable of organizing their own nucleolus. (iii) The heterozygous condition of most extra-NORs detected, as well as their presence in only some individuals of the populations analysed, indicate the instability of this phenomenon. However, contrary to other reports (Schubert & Wobus, 1985; Galetti et al., 1995), no sound evidence of intra-individual variation in the number and location of NORs was observed after applying CMA3-staining and/or rDNA-FISH. Furthermore, some extra-NORs could unambiguously be identified in several individuals of the same population (many of them were population-specific), which suggests their stable transmission across meiotic events and generations. Therefore, extra-NORs in brown trout, once originated, seem to integrate in a stable way into the karyotype, at least in the short term.
Several mechanisms have been invoked to explain the dispersion of rDNA loci from the ancestral pair in species with multichromosomal NOR distribution. The close association of NORs during nucleolus formation and the presence of heterochromatin in these regions could facilitate rearrangements between NOR-bearing chromosomes, as seem to be involved in the origin of specific NOR sites in the genus Salvelinus (Reed & Phillips, 1995). However, it is very unlikely that such rearrangements could have originated all the NOR-site variation detected in S. trutta without disturbing its specific karyotype.
It has been proposed that NOR regions could behave like mobile elements capable of spreading into the karyotype without altering the normal banding patterns (Schubert & Wobus, 1985; Galetti et al., 1995). This spreading could involve mobile elements associated with ribosomal genes (Jakubczak et al., 1992) or sequences dispersed throughout the genome, which could act as templates for amplification or recombination spots for rDNA loci (Maggini et al., 1991). In brown trout, the BglII element, a SINE-like sequence in the IGS close to the 18S RNA gene (Abuín et al., 1996; Morán et al., 1997), or the telomeric sequence (TTAGGG)n, which has been reported to be scattered through rDNA in different species (Reed & Phillips, 1995), including S. trutta (Abuín et al., 1996), could be responsible for the origin of extra-NORs. The exclusive telomeric location of active extra-NORs in our study supports this possibility.
All these sequences (BglII, telomeric consensus), however, are present both in the genome of standard and polymorphic individuals, and also in species which do not show such NOR-site variation. Therefore, they do not constitute per se an explanation for this phenomenon. A similar NOR dispersion to that described in Salvelinus and brown trout, has been recently detected in other salmonids of the subfamily Coregoninae (Jankun et al., unpublished data) and in one population of brown trout from Poland (Woznicki et al., 2000), which suggests its possible recurrence, even within this species. Taking one NOR-bearing pair as the plesiomorphic condition in this order, or two after the tetraploidization event, it looks as if NOR instability has appeared independently in both branches of Salmoniformes, or even in different areas of the distribution range of a single species. The karyotypic instability of this group related to its tetraploid origin, but also its large population subdivision and habitat fragmentation, could be additional factors to explain the recurrence of this phenomenon. These facts would facilitate reproductive isolation and also the consolidation of particular evolutionary events, like the polymorphism described.
Population analysis
The analysis of NOR-site variation in the populations sampled has shed some light about different evolutionary issues related to this polymorphism. Out of the four river drainages analysed in our study, the Miño basin was the most affected by the NOR-site polymorphism, especially around its central area. Although the neighbouring river basins (Umia and Tambre) also appeared to be affected, its intensity (nT, T, % atypical fish) was significantly much lower, all trout from the northernmost basin (Navia) being standard. In fact, we could detect a negative correlation between nT values and geographical distances to an ideal point located in the middle area of the Miño basin (rs=−0.64; P=0.002). These results suggest a single geographical origin for this NOR-site polymorphism, from which it would radiate to neighbouring areas. It is interesting to note that all extra-NORs described in the Umia or Tambre basins were also shared by populations from the Miño basin. The partial isolation of the Miño basin, located just at the southern limit of the anadromous form (Bouza et al., 1999), could have limited a higher dispersion of this phenomenon.
No polymorphic fish were detected in two samples from the Miño basin, Nogueira and Fondós. Fondós is a population fully isolated from the main watercourse by a waterfall which prevents the ascent of breeding trout. No field data are available about the putative isolation of Nogueira population. However, genetic distances calculated from isozyme data suggest some kind of isolation (Bouza et al., 1999). Deva and Requeixo, the two other populations well isolated from Miño, do contain polymorphic individuals, although their degree of polymorphism is amongst the lowest in this river basin. These results are in accordance with the lower genetic diversity usually observed in isolated populations with different genetic markers in brown trout (Bouza et al., 1999). The action of genetic drift on isolated populations is expected to lead to lower genetic diversity values.
Some information related to the temporal origin of this polymorphism also emerges from our data. Since some polymorphic individuals could be detected in isolated populations, the origin of this phenomenon should be previous to their isolation. In fact, the most highly differentiated population, Deva (D mean = 0.035), presented atypical individuals. This population showed a heterozygosity very close to non-isolated populations by using 33 allozyme loci, which suggests that no important bottlenecks have affected it (Bouza et al., 1999). Therefore, a gross estimation of the time of divergence (isolation) of this population could be obtained from Nei’s approach (Nei, 1987), indicating that this phenomenon could have appeared in brown trout from this area before 175 000 years ago. Of course, partial isolation, or human manipulation could disturb this estimate, which anyway should be taken with caution.
The distribution of allozyme variation in populations is usually explained as a counterbalance between migration and genetic drift (Chakraborty & Leimar, 1987). This is especially evident in Salmonids like S. trutta, with small effective population size (Jorde & Ryman, 1995), where a greater influence of genetic drift over genetic diversity and its distribution is expected. In our study, the distribution of allozyme and NOR-site variation within and among populations was globally very similar and nonsignificant (GST; Nei, 1977). Nevertheless, a different distribution of genetic variants was evident for both genetic markers. So, while isozyme diversity was mainly determined by differences in frequencies among populations, rather than by the presence of population-specific variants, NOR-site divergence among populations involved specific variants at low frequencies. Both factors counterbalance to render a similar GST estimate. In fact, genetic relationships among populations were very different when a detailed analysis was performed starting from genetic distances from all population pairs (D; Nei, 1978). Several explanations could account for the different genetic relationships observed with allozymes and NOR-site variation. The putative recent and single geographical origin of the NOR-site polymorphism could lead to a clinal distribution largely different to the isolation-by-distance relationships observed for allozymes. Also, the highly dispersed pattern detected for NOR-site variants seems to indicate that, whatever the mechanism of NOR jumping, it could be still active. If so, and assuming that extra-NORs are randomly dispersed to telomeric positions, transposition events would tend to increase the divergence among populations. Finally, the large excess of rRNA genes observed in our study (up to eightfold more rRNA genes than in standard ones; Castro et al., 1997), could generate important selective coefficients against high copy number, as suggested for higher eukaryotes (Lyckegaard & Clark, 1991).
In this work we have characterized a striking NOR-site polymorphism in S. trutta from a restricted area in North-western Spain. The results obtained confirm that extra-NORs are active and stable rDNA clusters always located at telomeric positions. The population analysis performed suggests a recent and single geographical origin of this polymorphism, and the possibility that, whatever the transposition phenomenon responsible, it could be still active.
References
Abuín, M., Clabby, C., Martínez, P. and Goswami, U. et al (1996). A NOR-associated repetitive element present in the genome of two Salmo species (Salmo salar and Salmo trutta). Genome, 39: 671–679.
Amemiya, C. T. and Gold, J. R. (1988). Chromosomal NORs as taxonomic and systematic characters in North American cyprinid fishes. Genetica, 76: 81–90.
Bouza, C., Arias, J., Castro, J. and Sánchez, L. et al. (1999). Genetic structure of brown trout, Salmo trutta L., at the southern limit of the distribution range of the anadromous form. Mol Ecol, 8: 1991–2001, 10.1046/j.1365-294x.1999.00794.x.
Castro, J., Rodríguez, S., Arias, J. and Sánchez, L. et al (1994). A population analysis of Robertsonian and Ag-NOR polymorphisms in brown trout (Salmo trutta). Theor Appl Genet, 89: 105–111.
Castro, J., Viñas, A., Sánchez, L. and Martínez, P. (1996). Characterization of an atypical NOR-site polymorphism in brown trout (Salmo trutta) with Ag- and CMA3-staining, and fluorescent in situ hybridization. Cytogenet Cell Genet, 75: 234–239.
Castro, J., de Lucchini, S., Nardi, I. and Sánchez, L. et al (1997). Molecular analysis of a NOR-site polymorphism in brown trout (Salmo trutta): organization of rDNA intergenic spacers. Genome, 40: 916–922.
Chakraborty, R. and Leimar, O. (1987). Genetic variation within a subdivided population. In: Ryman, N. and Utter, F. M. (eds). Population Genetics and Fishery Management, pp. 89–119. Washington Sea Grant Program, Washington University Press, Seattle.
Chakraborty, R., Haag, M., Ryman, N. and Stahl, G. (1982). Hierarchical gene diversity analysis and its application to trout population data. Hereditas, 97: 12–21.
de Lucchini, S., Nardi, I., Barsacchi, G. and Batistioni, R. et al (1993). Molecular cytogenetics of ribosomal (18S+28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome, 36: 762–763.
Dubcovsky, J. and Dvorak, J. (1995). Ribosomal RNA multigene loci: Nomads of the Triticeae genomes. Genetics, 140: 1367–1377.
Galetti, P. M., Mestriner, C. A., Monaco, P. J. and Rasch, E. M. (1995). Post-zygotic modifications and intra- and inter-individual nucleolar organizing regions variations in fish: report of a case involving Leptorinus friderici. Chrom Res, 3: 285–290.
Hall, K. J. and Parker, J. S. (1995). Stable chromosome fission associated with rDNA mobility. Chrom Res, 3: 417–422.
Howell, W. M. and Black, D. A. (1980). Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia, 36: 1014–1015.
Jakubczak, J. L., Zenni, M. K., Woodruff, R. C. and Eickbush, T. H. (1992). Turnover of R1 (type I) and R2 (type II) retrotransposable elements in the ribosomal DNA of Drosophila melanogaster. Genetics, 131: 129–142.
Jorde, P. E. and Ryman, N. (1995). Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics, 139: 1077–1090.
Lyckegaard, E. M. S. and Clark, A. G. (1991). Evolution of ribosomal RNA gene copy number on the sex chromosomes of Drosophila melanogaster. Mol Biol Evol, 8: 458–474.
Maggini, F. T., Cremonini, R., Zolfino, C. and Tucci, G. F. et al (1991). Structure and chromosome localization of DNA sequences related to ribosomal subrepeats in Vicia faba. Chromosoma, 100: 229–234.
Martínez, P., Viñas, A., Bouza, C. and Arias, J. et al. (1991). Cytogenetical characterization of hatchery stocks and natural populations of Sea and Brown Trout from Northwestern Spain. Heredity, 66: 9–17.
Martínez, P., Viñas, A., Bouza, C. and Castro, J. et al. (1993). Quantitative analysis of the variability of NOR region in Salmo trutta. Genome, 36: 1119–1123.
Morán, P., Reed, K. M., Pérez, J. and Oakley, T. et al. (1997). Physical localization and characterization of the BglI element in the genomes of Atlantic salmon (Salmo salar L.) and brown trout (S. trutta L.). Gene, 194: 9–18, 10.1016/s0378-1119(97)00093-0.
Moss, T. and Stefanovsky, V. Y. (1995). Promotion and regulation of ribosomal transcription by RNA polymerase I. Progr Nucl Acids, 50: 25–66.
Nei, M. (1977). F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet, 41: 225–233.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nei, M. (1987) Molecular Evolutionary Genetics. Columbia University Press, New York.
Phillips, R. B. and Ihssen, P. E. (1985). Chromosome banding in Salmonid fish: nucleolar organizer regions in Salmo and Salvelinus. Can J Genet Cytol, 27: 433–440.
Phillips, R. B., Zajicek, K. D. and Utter, F. M. (1986). Chromosome banding in Salmonid fishes: nucleolar organizer regions in Oncorhynchus. Can J Genet Cytol, 28: 502–510.
Phillips, R. B., Pleyte, K. A. and Hartley, S. E. (1988). Stock-specific differences in the number and chromosome positions of the nucleolar organizer regions in Arctic charr (Salvelinus alpinus). Cytogenet Cell Genet, 48: 9–12.
Reed, K. M. and Phillips, R. B. (1995). Molecular cytogenetic analysis of the double CMA3 chromosome of lake trout, Salvelinus namaycush. Cytogenet Cell Genet, 70: 104–107.
Saitou, N. and Nei, M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol, 4: 406–425.
Schmid, M., Feichtinger, W., Weimer, R. and Mais, C. et al (1995). Chromosome banding in Amphibia. XXI. Inversion polymorphism and multiple nucleolus organizer regions in Agalychnis callidryas (Anura, Hylidae). Cytogenet Cell Genet, 69: 18–26.
Schubert, I. and Wobus, U. (1985). In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma, 92: 143–148.
Schweizer, D. (1979). Fluorescent chromosome banding in plants: applications, mechanisms and implications for chromosome structure. In: Davies, D. R. and Hopwood, R. A. (eds). The Plant Genome, pp. 61–72. Proc. 4th John Innes Symposium, John Innes Charity, Norwich.
Suzuki, H., Kurihara, Y., Kanemisha, Y. and Moriwaki, K. (1990). Variation in the distribution of silver-staining nucleolar organizer regions on the chromosomes of the wild mouse Mus musculus. Mol Biol Evol, 7: 271–282.
Volleth, M. (1987). Differences in the location of nucleolus organizer regions in European vespertilionid bats. Cytogenet Cell Genet, 44: 186–197.
Woznicki, P., Sánchez, L., Martínez, P. and Pardo, B. G. et al (2000). A population analysis of the structure and variability of NOR in Salmo trutta by Ag, CMA3 and ISH. Genetica, 108: 113–118, 10.1023/a:1004055125295.
Zhuo, L., Reed, K. M. and Phillips, R. B. (1995). Hypervariability of ribosomal DNA at multiple chromosomal sites in lake trout (Salvelinus namaycush). Genome, 38: 487–496.
Acknowledgements
This study was supported by a grant from Xunta de Galicia (XUGA26102B97). The authors want to thank A. Caballero, P. M. Galetti, and C. Bouza for comments on the manuscript. They have aided in the improvement of the manuscript. The computer programs NEGST and DISPAN were kindly supplied by N. Ryman (Stockholm University, Sweden) and M. Nei (Pennsylvania State University, U.S.A.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Castro, J., Rodríguez, S., Pardo, B. et al. Population analysis of an unusual NOR-site polymorphism in brown trout (Salmo trutta L.). Heredity 86, 291–302 (2001). https://doi.org/10.1046/j.1365-2540.2001.00834.x
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.2001.00834.x
Keywords
This article is cited by
-
Karyotypic variation in the stingless bee Trigona spinipes (Hymenoptera: Apidae: Meliponini) from different geographical regions of Brazil
Apidologie (2021)
-
How dynamic could be the 45S rDNA cistron? An intriguing variability in a grasshopper species revealed by integration of chromosomal and genomic data
Chromosoma (2019)
-
Evolutionary trends in animal ribosomal DNA loci: introduction to a new online database
Chromosoma (2018)
-
Molecular cytogenetic analysis reveals the existence of two independent neo-XY sex chromosome systems in Anatolian Pamphagidae grasshoppers
BMC Evolutionary Biology (2017)
-
Chromosomal characteristics and distribution of rDNA sequences in the brook trout Salvelinus fontinalis (Mitchill, 1814)
Genetica (2015)