Abstract
Twenty-six old French six-rowed winter barley (Hordeum vulgare L.) varieties were characterized for their reaction against barley mild mosaic virus (BaMMV). The genetic diversity of these varieties and two recent barley varieties was assessed using molecular, biochemical and morphological data. Seven old varieties were fully resistant to BaMMV. A higher differentiation level between varieties was observed by using DNA molecular markers compared to biochemical and morphological ones. Correspondence analysis using all markers showed that DNA molecular data could fully discriminate between all varieties, whereas biochemical and morphological markers were not able to achieve a complete discrimination. The dendrogram clustering computed with the DNA marker dissimilarity index showed two main groups. The first group included the seven varieties resistant to the BaMMV, whereas the second contained susceptible varieties. The relationships between these varieties, their diversity level, and their characterization are discussed. We infer that the seven BaMMV-resistant varieties have a common origin.
Similar content being viewed by others
Introduction
Old French barley varieties which are derived directly from landraces, or have such material in their ancestry, are maintained at INRA. They have been studied for their morphological and agronomic diversity in a national network (Koenig et al., 1991), and represent a potential source of genetic diversity which may be valuable in the future. It has been shown that some of them carried resistance genes to the Barley Mosaic complex (Le Gouis & Hariri, 1996). The characterization of these genetic resources, and the assessment of their diversity and phylogenetic relationships, will provide useful information for their more efficient exploitation in breeding programmes.
Botanical and agronomic traits both have limitations in the discrimination of barley varieties, and it is generally difficult to identify barley cultivars on the basis of seed morphology (Hoffman & Bregitzer, 1996). Other markers such as isozymes, proteins and hordeins have been used successfully in the analysis and identification of varieties (Hoffman & Goates, 1990; Konishi & Matsuura, 1991; Bernardo et al., 1997). Although isozymes are fast and easy to use, their relatively low number and level of variability limit their use in genetic diversity studies (Le Gouis et al., 1995).
Molecular markers, such as RFLP, AFLP, STS and microsatellites, present the most powerful way to access genetic diversity, to undertake genetic mapping and to identify different varieties (cf. Graner et al., 1991; Saghai-Maroof et al., 1994; Becker et al., 1995). In Hordeum species these markers have made possible the characterization of different cultivars, the understanding of phylogenetic relationships, and genetic mapping (Hoffman & Bregitzer, 1996; Marillia & Scoles, 1996; Dahleen et al., 1997; Ordon et al., 1997).
The aim of this work was to study the genetic diversity of old French six-rowed barley varieties in relation to resistance to BaMMV with the help of morphological markers, isozymes, RAPDs and microsatellites.
Materials and methods
Plant materials and BaMMV test
Twenty-six old French winter barley varieties (Table 1) kept in the INRA collection at Clermont-Ferrand were used in this study. They represent most of genetic resources in winter barley of French origin. Seven to 34 plants of each genotype were mechanically inoculated with BaMMV at the three-leaf stage. The DAS-ELISA test was carried out three weeks after the inoculation to assess resistance or susceptibility. Two widely grown modern cultivars were added in the study: cv. Plaisant which is susceptible to BaMMV, and cv. Express which is resistant.
Botanical and morphological markers
Eight characters were measured on plants sown in a nursery (three rows of 25 plants) during two growing seasons at INRA Clermont-Ferrand: pilosity of kernel furrow (hairy or glabrous), pilosity of the rachilla (curly-haired or erect-haired), glume size (small, medium or large), number of spikelets (low, intermediate or high), spike compactness (lax, half-lax, half-dense or dense), thousand kernel weight (low, intermediate or high), resistance to powdery mildew (Erysiphe graminis f. sp. hordei) (low, intermediate or high), and lodging resistance (low, intermediate or high).
Isozyme markers
Equal amounts of leaf tissue from 10-day-old plantlets were crushed in a buffer solution (Ostergaard et al., 1985). Electrophoresis was performed in 12% starch gels according to Trigui et al. (1986). Three plants of each of the 28 varieties were analysed for the following isozyme systems: esterase (Est), NADH dehydrogenase (Ndh), glutamic-oxaloacetic transaminase (Got) and 6-phosphogluconate dehydrogenase (Pgd). Staining procedures and nomenclature designation of the loci and alleles followed Nielsen & Johansen (1986) and Hvid & Nielsen (1977). A supplementary allele Hg (reference variety ‘Hâtif de Grignon’) was found in addition to the common allele Ma (reference variety ‘Mammut’) at the Ndh2 locus.
DNA markers
Seeds from the 28 varieties were germinated in Petri dishes for seven days. Young seedlings were collected and kept frozen at −80°C. In order to assess uniformity of accessions, we individually extracted five seedlings of 10 varieties and performed RAPDs with primer G11. Two repetitions for each amplification were carried out with a one-week interval to test uniformity of RAPD patterns. For primer Z04 A, amplifications were performed from DNA individually extracted from two plants per genotype. In each case, amplification patterns of individual plants or bulked plants for the same genotype remained identical. Then for each variety five seedlings were bulked and DNA was extracted according to Plomion et al. (1995). The extracts were treated with RNAase. RAPD reactions were performed with the following 16, 10-base primers (Operon Technologies Inc., Alameda, USA): AC09 (5′-AGAGCGTACC-3′), AD15 (5′-TTTGCCCCGT-3′), G11 (5′-TGCCCGTCGT-3′), AB06 (5′-GTGGCTTGGA-3′), AC02 (5′-GTCGTCGTCT-3′), AA19 (5′-TGAGGCGTGT-3′), AC05 (5′-GTTAGTGCGG-3′), AB05 (5′-CCCGAAGCGA-3′), G04 (5′-AGCGTGTCTG-3′), A04 (5′-AATCGGGCTG-3′), G06 (5′-GTGCCTAACC-3′), F01 (5′-ACGGATCCTG-3′), C04 (5′-CCGCATCTAC-3′), J16 (5′-CTGCTTAGGG-3′), N07 (5′-CAGCCCAGAG-3′) and O19 (5′-GGTGCACGTT-3′). The reaction mixture (20 μL) contained 15 ng of DNA, 100 μM of each dNTP, 2× buffer (Appligene Oncor), 0.3 μM primer, 0.5 U of Taq DNA Polymerase (Appligene Oncor). Amplifications were carried out using a Genius thermocycler (Techne, Cambridge, UK). A first denaturation step (94°C for 4 min) was followed by 45 cycles at: 94°C for 1 min, 40°C for 1 min and 72°C for 2 min. A Perkin Elmer 2400 thermocycler was used for an 11-base primer Z04A (5′-AGGCTGTGCTA-3′) developed by Schiemann et al. (1997). The reaction mixture (20 μL) was composed of 1.5 U of AmpliTaq DNA Polymerase Stoffel fragment (Perkin-Elmer), 2× Stoffel buffer, 400 μM each of dNTP, 6 mM MgCl2 and 0.3 μM primer. The amplification was conducted at 94°C for 4 min for one cycle; and at 94°C for 1 min, 44°C for 1 min, 72°C for 2 min (the polymerization time was extended 3 s per cycle) for 45 cycles. Amplification products were separated in 1.8% agarose gels, and detected by ethidium bromide staining.
Two microsatellites, HVM3 and HVM70, were also used (Liu et al., 1996). The reaction volume (25 μL) contained 200 μM of each dNTP, 0.4 pM of each primer, 20 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2, 0.9 U of Taq DNA polymerase (GIBCO BRL, Life Technologies) and 7 ng of DNA. PCR amplification was performed according to Liu et al. (1996). Amplification products were separated in 6% polyacrylamide sequencing gels and silver stained.
Finally, the two primers (5′-CAGGAGCTGGTGAAATAGTGCCT-3′ and 5′-TTAAAGGGCTCCGTGAAGC-3′) for the YLM marker of the Yd2 resistance gene to Barley Yellow Dwarf Virus, were used and scored according to Paltridge et al. (1998).
Statistical analysis
Statistical analyses were performed on a binary matrix. Each variable RAPD band was considered as a locus so that every locus had two alleles. For correspondence analysis, only polymorphic variables were kept. Dissimilarity indices were computed between all pairs of varieties, using the ‘Euclidean’ (Euclidean distances are root sum-of-squares of differences) method (S-PLUS, 1991). Then the dendrograms were plotted using the dissimilarity data, according to the average method of clustering (single-linkage) with the SPLUS system version 3.0 (1991). Data were first processed separately for molecular, biochemical and agronomic markers; the analysis was then performed on all markers simultaneously.
Results and discussion
Table 1 shows the result of mechanical inoculation with BaMMV. No infected plants were detected for seven cultivars: Comte de Serre, Demi-Hâtif des Tourettes, Escourgeon de Champagne, Escourgeon de la Marne, Hâtif de Grignon, Ile de Ré and Superchampenois. Resistance in the field to BaMMV and BaYMV has also been reported by Bensalah et al. (1998) for all these cultivars except Demi-Hâtif des Tourettes. For the other 19 genotypes, infection rates (number of infected plants/number of tested plants) were between 20 and 100%. Repeated infections for genotypes with a low number of infected plants (Ardennes, Demi-Hâtif Lafite, Dunois, Hâtif Bonté and Précoce Lafite) gave consistent results. Residual heterozygosity can not be eliminated as an explanation of this intermediate behaviour. Partial resistance to mechanical inoculation has already been reported (Proeseler et al., 1988; Graner et al., 1995) with lower infection rates and virus concentrations and longer incubation periods.
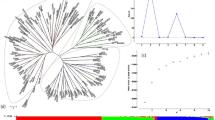
Analysis of morphological and botanical characters showed that the specificity of these markers for genotype discrimination was limited. For example, the 28 varieties could be classified into three classes when using a combination of two botanical characters. Six varieties (Dunois, Escourgeon de la Marne, Escourgeon du Nord N°5, Hâtif de Grignon, Hexagonal de Wagnonville and Précoce Lafite) had a glabrous furrow kernel and an erect-haired rachilla. Eight varieties (Demi-Hâtif des Tourettes, Dijon, Escourgeon de Champagne, Escourgeon de Valence, Escourgeon Extra-Hâtif, Ile de Ré and Superchampenois) were characterized by a glabrous furrow and a curly-haired rachilla. A hairy furrow and an erect-haired rachilla distinguished the last 14 varieties. No variety had both a hairy furrow and a curly-haired rachilla. Individual identification of the varieties was not possible using all the morphological traits. Correspondence analysis with 23 morphological characters showed that only the variety Dunois was separated (Fig. 1a). This variety is now thought to have originated from a mutation within the German cultivar Probstdorf. A dendrogram plotted with the morphological data (Fig. 2a) showed more than four individual clusters.
Correspondence analysis: (a) with morphological data; (b) isozyme data; (c) molecular markers; (d) all markers. Resistant varieties are indicated by open circles. For abbreviations see Table 1.
Dendrograms computed by average clustering: (a) with morphological data; (b) isozyme data; (c) molecular markers; (d) all markers. Scales are the percentage of similarity. Resistant varieties are indicated by *. For abbreviations see Table 1.
Among a total of nine isozyme loci analysed, seven were variable: Est1 (three alleles: Ca, Pr, Al), Est4 (two alleles: Su, At), Est5 (three alleles: Pi, Te, mi), Est9 (two alleles: ne, Fl), Ndh1 (two alleles: Je, An), Ndh2 (two alleles: Hg, Mu), Pgd2 (two alleles: Ps, Tn). Two loci were monomorphic: Est2 (allele Fr) and Got3. Eighteen alleles were detected and the mean number of alleles per locus was 2.0. The first plane of the correspondence analysis showed an enlarged distribution of the varieties where only Dijon was isolated from the other varieties (Fig. 1b). Cluster analyses displayed two major groups where the varieties were dispersed without any apparent biological relationships (Fig. 2b). Some varieties were grouped at the same level of similarity because of the very low isozyme diversity.
The number of bands per RAPD primer varied from 10 to 15. In total 193 bands were obtained using 17 primers; and 81 variable bands, considered here to correspond to 81 loci, were scored (45%). Nine varieties were distinguished on the basis of the presence or absence of only one specific band. Fifteen other varieties were identified on the basis of presence and/or absence of two or three bands. Finally, four varieties needed more than three bands to be differentiated.
Seven and three alleles were scored for HVM3 and HVM70, respectively. For HVM3, allele no. 3 was present in 11 varieties (39%) and allele no. 4 in 10 varieties (35%). Allele no. 1 of HVM70 was present in 23 varieties (82%). YLM amplification products revealed two bands. Six cultivars (Comte de Serre, Escourgeon de la Marne, Escourgeon Extra-Hâtif, Hâtif de Grignon, Hâtif du Douaisis, Superchampenois and Express) tested positively to the YLM-S band (Paltridge et al., 1998). The correspondence analysis using variable bands of all DNA molecular markers showed two groups (Fig. 1c). The first one contained Comte de Serre, Demi-Hâtif des Tourettes, Escourgeon de Champagne, Escourgeon de la Marne, Hâtif de Grignon, Ile de Ré, Superchampenois and Plaisant. Except for the modern cultivar Plaisant, all were resistant to BaMMV (Table 1). The dendrogram plotted with these data also showed a group including the same seven resistant varieties (Fig. 2c), whereas Express and Plaisant were grouped together. This may be because they both share Ager as a common ancestor. It can also be noticed that four out of five cultivars which showed intermediate infection rates (Ardennes, Demi-Hâtif Lafite, Dunois and Précoce Lafite) are grouped together.
In the last analysis, a correspondence analysis was performed on data from all markers (162 RAPD alleles at 81 loci, 10 microsatellites alleles at two loci, two YLM alleles at one locus, 16 isozyme alleles at eight loci and 23 morphological characters) and the same two major groups were detected (Fig. 1d). Plaisant was now, however, in the same group as all the other susceptible old French barley varieties. The first two axes represented 23.8% of the total inertia. No additional grouping could be detected along the third (8.4% of total inertia) or fourth axis (7.7%).
The genetic determination of resistance to BaMMV of the seven cultivars is not fully known. Le Gouis & Hariri (1996) have shown that Comte de Serre, Escourgeon de la Marne and Superchampenois carried two resistance genes. One of these was allelic to ym4 as no segregation was observed in crosses with Express. Ordon et al. (1995) have shown that the OPZ04H660 marker was tightly linked to ym4 (0.7 cM). Furthermore, they showed that this marker could discriminate perfectly between resistant and susceptible commercial barley cultivars. We tested the more specific Z04A primer developed by Schiemann et al. (1997). Although this marker could logically differentiate Plaisant and Express, all the old French barley cultivars carried the 660 bp band expected to be characteristic of susceptible varieties. The ym4 gene is thought to come from the landrace Ragusa (Huth, 1985). This landrace was brought back from south Dalmatia in the Ragusa/Dubrovnik region in the late 1920s (von Aufhammer, 1982) and was later extensively used in crosses because of its powdery mildew resistance gene. Nothing is known about the possible relationships between French and Dalmatian barley populations. A different origin for the ym4 gene may explain why the 660 bp band could not discriminate between susceptible and resistant old French barley varieties. Another explanation could be that resistant old French barleys carry the ym5 gene which is likely to be allelic to ym4, but conferring a wider resistance. This would explain why these three cultivars are resistant to the second strain of BaYMV (Bensalah et al., 1998). The second gene carried by Comte de Serre, Escourgeon de la Marne and Superchampenois is not known with certainty. In the correspondence analysis with all markers, the susceptible and resistant genotypes were separated along the first axis, which represented 14.2% of total inertia. The maximum contribution to this axis was observed for the AC02H1200, AC02H1100, AA19H1600, AA19H1200, AB05H950, G06H680 and N07H550. Variable bands for AA19, AB05 and N07 (Dahleen et al., 1997), and G06 (Bauer et al., 1997) were mapped in the centromeric region of chromosome 4H. This information suggests that ym11 may be the second resistance gene as this gene was localized in this region. Crosses with Russia 57, which carries ym11 (Bauer et al., 1997), are needed to confirm this hypothesis. The resistance of the four other old French barley varieties has not yet been studied. Genetic similarity was found between all the resistant cultivars as shown in Figs 1 and 2. The same result was obtained when the markers mapped on chromosome 4H were removed (data not shown). This suggests that these seven cultivars are related and their resistance genes may be identical.
The origin of all the old French barley varieties is not fully known (Table 1). Comte de Serre is thought to come from the Paris Region. Its similarity with Superchampenois or Escourgeon de la Marne (Figs 1 and 2) suggests a common origin. For Carré d’hiver the lowest dissimilarity index was obtained with Escourgeon de Beauce, whereas for Orge d’hiver à six-rangs it was with Dijon. The geographical origin and the genetic similarity as measured by the markers we used did not appear to be correlated. The three genotypes from Charente Maritime (Demi-Hâtif des Tourettes, Hâtif de Grignon and Ile de Ré) had low dissimilarity indices. The same was true for the three cultivars from Marne (Escourgeon de Champagne, Escourgeon de la Marne and Superchampenois). The two genotypes from Vendée (Escourgeon de Vendée and Escourgeon Extra-Hâtif) or the genotypes from the North (Nord) were more dissimilar.
References
Bauer, E., Weyen, J., Schiemann, A., Graner, A. and Ordon, F. (1997). Molecular mapping of novel resistance genes against Barley Mild Mosaic Virus (BaMMV). Theor Appl Genet. 95: 1263–1269.
Becker, J., Vos, P., Kuiper, M., Salamini, F. and Heun, M. (1995). Combined mapping of AFLP and RFLP markers in barley. Mol Gen Genet. 249: 65–73.
Bensalah, B., Froidmont, F. and Jacquemin, J. -M. (1998). Screening barley germplasm for resistance to a new isolate of barley yellow mosaic virus (BaYMV). Agronomie. 18: 71–78.
Bernardo, A., Luque, A., Cuadrado, A., Negro, A., Jouve, N. and Soler, C. (1997). The assessment of genetic variation in Spanish primitive cultivars of barley, Hordeum vulgare L., by a combination of isozymes and hordeins. Gen Res Crop Evol. 44: 217–226.
Dahleen, L. S., Hoffman, D. L., Dohrmann, J., Gruber, R. and Franckowiak, J. (1997). Use of a subset of doubled-haploid lines for RAPD interval mapping in barley. Genome. 40: 626–632.
Graner, A., Jahoor, A., Schondelmaier, J., Siedler, H., Pillen, K., Fischbeck, G. et al (1991). Construction of an RFLP map of barley. Theor Appl Genet. 83: 250–256.
Graner, A., Bauer, E., Kellermann, A., Proeseler, G., Wenzel, G. and Ordon, F. (1995). RFLP analysis of resistance to the barley yellow mosaic virus complex. Agronomie. 15: 475–479.
Hoffman, D. L. and Bregitzer, P. (1996). Identification of reproducible PCR-RAPD markers that enable the identification of closely related six-rowed malting barley (Hordeum vulgare L.) cultivars. J Am Soc Brew Chem. 54: 172–176.
Hoffman, D. L. and Goates, B. J. (1990). Polymorphism and genetic basis of β-N-acetylglucosaminidase, hexokinase, and β-D-galactosidase in barley (Hordeum spp.). Biochem Genet. 28: 487–493.
Huth, W. (1985). Versuche zur Virusdiagnose und Resistenzträgererstellung in Gerste gegen Barley Yellow Mosaic Virus (BaYMV). Vortr Pflanz. 9: 107–120.
Hvid, S. and Nielsen, G. (1977). Esterase isozyme variants in barley. Hereditas. 87: 155–162.
Koenig, J., Letarga-Leblanc, A., Jestin, L., Legouis, J. and Bouguennec, A. (1991). Cereal genetic resources networks in France. In: Van Hintum, T. J. L., Frese, L. and Perret, P. M. (eds) Searching for New Concepts for Collaborative Genetics Resource Management, pp. 111–115. Eucarpia/IBPGR symposium, Wageningen, Netherlands.
Konishi, T. and Matsuura, S. (1991). Geographic differentiation in isozyme genotypes of Himalayan barley (Hordeum vulgare). Genome. 34: 704–709.
Legouis, J. and Hariri, D. (1996). Characterisation of the resistance of old winter barley (Hordeum vulgare L.) French cultivars to barley mosaic viruses. In: Proceedings of the V International Oat Conference and VII International Barley Genetics Symposium, Saskatoon, Saskatchewan, Canada, pp. 746–748.
Legouis, J., Erdogan, M., Friedt, W. and Ordon, F. (1995). Potential and limitations of isozymes for chromosomal localization of resistance genes against barley mild mosaic virus (BaMMV). Euphytica. 82: 25–30.
Liu, Z. W., Biyashev, R. M. and Saghai-Maroof, M. A. (1996). Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor Appl Genet. 93: 869–876.
Marillia, E. F. and Scoles, G. J. (1996). The use of RAPD markers in Hordeum phylogeny. Genome. 39: 646–654.
Nielsen, G. and Johansen, H. B. (1986). Proposal for the identification of barley varieties based on the genotypes for 2 hordeins and 39 isozyme loci of 47 reference varieties. Euphytica. 35: 717–728.
Ordon, F., Bauer, E., Friedt, W. and Graner, A. (1995). Marker-based selection for the ym4 BaMMV-resistance gene in barley using RAPDs. Agronomie. 15: 481–485.
Ordon, F., Schiemann, A. and Friedt, W. (1997). Assessment of the genetic relatedness of barley accessions (Hordeum vulgare s.l.) resistant to soil-borne mosaic-inducing viruses (BaMMV, BaYMV, BaYMV-2) using RAPDs. Theor Appl Genet. 94: 325–330.
Ostergaard, H., Nielsen, G. and Johansen, H. (1985). Genetic variation in cultivars of diploid ryegrass, Lolium perenne and L. multiflorum, at five enzyme systems. Theor Appl Genet. 69: 409–421.
Paltridge, N. G., Collins, N. C., Bendahmane, A. and Symons, R. H. (1998). Development of YLM, a codominant PCR marker closely linked to the Yd2 gene for resistance to barley yellow dwarf disease. Theor Appl Genet. 96: 1170–1177.
Plomion, C., Bahrman, N., Durel, C. -E. and O’malley, D. M. (1995). Genomic mapping in Pinus pinaster (maritime pine) using RAPD and protein markers. Heredity. 74: 661–668.
Proeseler, G., Reichenbächer, D. and Urban, M. (1988). Quantitative Resistenz der Wintergerste gegen Gerstengelbmosaic-Virus (barley yellow mosaic virus). Arch Phytopathol Pflanz, Berlin. 24: 441–443.
Saghai-Maroof, M. A., Biyashev, R. M., Yang, G. P., Zhang, Q. and Allard, R. W. (1994). Extraordinarily polymorphic microsatellite DNA in barley: species diversity, chromosomal locations, and population dynamics. Proc Natl Acad Sci USA. 91: 5466–5470.
Schiemann, A., Graner, A., Friedt, W. and Ordon, F. (1997). Specificity enhancement of a RAPD marker linked to the BaMMV/BaYMV resistance gene ym4 by randomly added bases. Barley Genet Newsl. 27: 63–65.
S-PLUS (1991). S-Plus user’s manual, version 3.0., Statistical Sciences, Inc., Seattle, WA.
Trigui, N., Sandmeier, M., Salanoubat, M. and Pernes, J. (1986). Utilisation des données enzymatiques et morphologiques pour l’étude des populations et de la domestication des plantes. I. Séparation et identification génétique d’isozymes chez le mil (Pennisetum typhoides Bur. Stapf and Hubb.). Agronomie. 6: 779–788.
Vonaufhammer, G. (1982). Zur Abstammung von Wintergersten-sorten-ahnentafel. Bayer-Landw Jb. 59: 536–544.
Acknowledgements
This research was supported in part by a grant of the International Bureau for Genetic Resources. We thank N. Paltridge for PCR markers for Yd2. We thank two anonymous referees for their helpful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bahrman, N., Le Gouis, J., Hariri, D. et al. Genetic diversity of old French six-rowed winter barley varieties assessed with molecular, biochemical and morphological markers and its relation to BaMMV resistance. Heredity 83, 568–574 (1999). https://doi.org/10.1038/sj.hdy.6886140
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6886140