Abstract
Nine B chromosome variants, from seven different populations of Eyprepocnemis plorans collected at four localities in Spain and three in Morocco, have been shown to be mainly composed of two DNA sequences, i.e. a 180-bp tandem repeat and ribosomal DNA. B types, however, differ in the relative amounts of the two sequences. The most widespread one (B1) bears about the same amount of rDNA and 180 bp repeat, but three other variants that have reached a polymorphism by replacing B1 in smaller areas (B2, B5 and B24) carry a conspicuously larger amount of the 180 bp repeat. In Morocco, the most widespread B variant is also B1, and a rare variant that appeared in a single individual is also built with the same two DNA sequences. All these data point to a common origin for these B chromosomes, with B1 probably being the original one. The origin of the different B types and the possible relationship of the relative amount of 180 bp DNA repeat with B drive are discussed.
Similar content being viewed by others
Introduction
Supernumerary, accessory or B chromosomes are dispensable extra chromosomes that are present in some individuals from some populations in many plant and animal species. Their main characteristic is that they do not recombine with the standard chromosomes and thus follow their own evolutionary pathway (J. P. M. Camacho and J. S. Parker, in Beukeboom, 1994). Intensive investigation during the last decade has shown that B chromosomes are mainly composed of repetitive DNA. B-specific DNA sequences have been found in some cases, but in others all B sequences were homologous to DNA sequences in the standard chromosomes (for review, see Beukeboom, 1994 and Camacho et al., 1999). A simple inference in the case of shared sequences is that Bs are derived from the standard chromosomes of the same species (intraspecific hypothesis of B origin). B-specific DNA sequences might have arisen by either rapid evolution of the B-DNA, which thus loses its homology with the A chromosome sequences from which it was derived (intraspecific hypothesis), or else from interspecific hybridization (interspecific hypothesis). In the latter case, B-specific sequences have been found showing homology with DNA sequences present in the A chromosomes of a closely related species (John et al., 1991; Eickbush et al., 1992).
The grasshopper Eyprepocnemis plorans shows a highly variable (more than 40 different B variants have hitherto been reported; López-León et al., 1993) and widespread (Bs are present in most field populations hitherto analysed) B chromosome polymorphism along the Mediterranean and Atlantic coasts of the Iberian Peninsula and North Africa. It provides the most complete example of selfish B chromosome evolution identified to date, showing B neutralization by the host genome and the regeneration of the polymorphism by the replacement of the neutralized B by a new selfish B showing drive (Camacho et al., 1997a,b; Zurita et al., 1998). Previous investigation showed that B2, the most common B variant in natural populations from the Granada province, is mainly composed of two DNA sequences, a 180-bp DNA repeat and ribosomal DNA (López-León et al., 1994). Sequence analysis indicated that the 180-bp tandem repeat is very unlikely to encode a protein (López-León et al., 1995). Likewise, the rDNA is usually inactive, although it conserves the capability for expression, as shown by an exceptional male with the B2 fused to the longest autosome and showing an active nucleolus organizing region (NOR) in the distal region of the B (Cabrero et al., 1987), thus suggesting low or no sequence divergence in the B rDNA from that of standard chromosomes.
In the present paper, we present evidence that the nine analysed B chromosome variants in E. plorans, collected in geographically distant populations from Spain and Morocco, are composed primarily of these two DNA sequences, suggesting a common origin for all these Bs.
Materials and methods
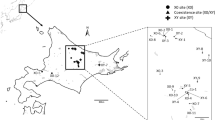
Adult males of E. plorans were collected at four Spanish localities: Mundo river (Albacete), Salobreña (Granada), Fuengirola (Málaga) and Torrox (Málaga); and three Moroccan populations: SO.DE.A., Ain l’abid and Smir. Testes were fixed in freshly prepared 3:1 ethanol:acetic acid and stored at 4°C. Chromosomal preparations were made by squashing two testis follicles in 50% acetic acid. After 10 min, the coverslip was removed by immersing the preparation in liquid nitrogen for a few seconds and then the slides were air-dried. Some of the slides were subjected to the C-banding technique described in Camacho et al. (1991) in order to characterize the B chromosome types, and the remaining slides were used for fluorescent in situ hybridization (FISH).
To eliminate the large amount of cytoplasm present in grasshopper meiocytes, in order to allow probe access to chromosomal DNA, each preparation was incubated in 150 μL per slide of 50 μL/mL pepsin in 0.01N HCl at 37°C in a humid chamber for 15 min and washed in distilled water. Preparations were then dehydrated in an ethanol series, air-dried and placed in an oven at 37°C overnight. Slides were then subjected to the double FISH technique described in López-León et al. (1994), with two probes: (i) pTa71 from wheat (Triticum aestivum), which is a complete 9 kb rDNA gene unit containing the 5.8S, 18S and 26S genes and the intergenic spacer (Gerlach & Bedbrook, 1979), kindly provided by R. B. Flavell and M. O’Dell, and (ii) pEpD15, which is a 180-bp DraI fragment of repetitive DNA from E. plorans (see López-León et al., 1994, 1995). Both pTa71 and pEpD15 were labelled by nick translation with FluoroGreen (fluorescein-11-dUTP) and FluoroRed (rhodamine-11-dUTP) from Amersham, respectively. Preparations were examined with a Nikon epifluorescence microscope and photographs were taken on Fujichrome 400 Provia colour film. Slides were digitized with a Hewlett Packard Photo Smart scanner and the figures were composed with Adobe Photoshop and Microsoft Word 97.
Results
A total of nine B chromosome variants were analysed, six from Spanish populations and three from Moroccan ones. The Spanish variants included the most widespread one, B1, which is present throughout the whole Mediterranean and southern Atlantic coasts of the Iberian Peninsula, and hence is considered to be the original B (Henriques-Gil et al., 1984); this variant was collected at the river Mundo (Fig. 1). The remaining five Bs from Spanish populations were B2 (Fig. 1b) and a large metacentric B (Fig. 1c), both from Salobreña, B24 (Fig. 1d) and a small B (Fig. 1e) from Torrox, and B5 (Fig. 1f) from Fuengirola. Three of these B variants, i.e. B2, B24 and B5, were the most frequent B type in their respective localities, where they have replaced the former predominant B (B1 for B2 and B5, and B2 for B24) (Henriques-Gil & Arana, 1990; Zurita et al., 1998).
Double fluorescent in situ hybridization (FISH) on nine B chromosomes in Spanish and Moroccan populations of the grasshopper Eyprepocnemis plorans. Ribosomal DNA is shown in green and the 180 bp DNA repeat in red. The B chromosomes from Spain were found at (a) Mundo river (B1), (b) Salobreña (B2), (c) Salobreña (iso-B2), (d) Torrox (B24), (e) Torrox (small B) and (f) Fuengirola (B5). The B chromosomes from Morocco were found at (a) SO.DE.A. (B1), (b) Ain l’abid (B1) and (c) Smir (large B).
In Morocco, the predominant B in all populations hitherto analysed shows a C-banding pattern similar to the Spanish B1 (Bakkali et al., 1999). Representatives of this B type are shown in Fig. 1(a) (SO.DE.A.) and Fig. 1(b) (Ain l’abid). Fig. 1(c) shows a rare B variant that was present in a single individual from Smir.
All Bs were acro-telocentric except the large metacentric one from Salobreña (Fig. 1c, upper row). The principal B types possess a small short arm that must contain DNA different from the two probes assayed, because double FISH (Fig. 1) does not reveal it. The following results will therefore refer to the long arm of the acro-telocentric Bs.
The most remarkable fact shown by Fig. 1 is that the molecular composition of all these B variants is rather similar, all of them being essentially composed of the two DNA sequences assayed, and differing only in the relative amount of the 180 bp DNA-repeat and the rDNA. B1 from Mundo river (Fig. 1a) possessed almost identical amounts of both DNA sequences, and the same ratio was present in the predominant B in Morocco (Fig. 1a, b) which should also be considered as the B1 type. B2 and B5, however, carried a 180-bp DNA-repeat amount about double that of rDNA (Fig. 1b, upper row1 and), and B24 carried a 180-bp DNA-repeat amount about five times that of rDNA (Fig. 1d). Finally, the three rare B types, i.e. the large metacentric B from Salobreña (Fig. 1c, upper row), the small B from Torrox (Fig. 1e) and the large B from Smir (Fig. 1c, lower row) were all also mainly composed of the same two DNA sequences as the principal B types.
Discussion
The extremely high variation of the B chromosome polymorphism in E. plorans, with more than 40 different B types differentiated in terms of size, morphology and C-banding pattern, seems to be an inherent property of these Bs (López-León et al., 1993). The present results show that all B variants analysed are mainly composed of the same two DNA sequences (rDNA and a 180-bp tandem repeat), although at different relative amounts. This suggests a common origin for these B types from an original B that most likely was B1, given its much broader geographical distribution in both the Iberian Peninsula and Morocco (Henriques-Gil et al., 1984). It is remarkable that the order of the two DNA sequences with respect to the centromere has been conserved in all B variants, consistent with the order in the presumed ancestral chromosome from the A set, the X chromosome (López-León et al., 1994). This suggests that the 180 bp DNA repeat might be involved in centromere function, and the absence of this DNA in the centromeric region of some A chromosomes (López-León et al., 1994) does not preclude this possibility because such a function is not necessarily sequence-dependent, as is illustrated by the lack of a repeat DNA sequence shared by centromeres of different organisms, or by the fact that in Drosophila melanogaster no single sequence is shared by all centromeres (Karpen & Allshire, 1997).
The processes by which B chromosome variation has been achieved in E. plorans, from the ancestral B1, are mainly the differential amplification or deletion of these two DNA sequences, by means of unequal crossover. For example, B2 and B5 are simply B1 chromosomes that have lost part of their rDNA and have gained some amount of the 180 bp repeat. Likewise, B24 is a B2 that has increased 180 bp repeat sequences and lost rDNA. The small B found in the Torrox population, where B24 has recently replaced B2 (Zurita et al., 1998), is very interesting because it could have had a concomitant origin with B24. An unequal crossover between two B2 chromosomes could have simultaneously yielded B24 and the small B (Fig. 2). The alternative hypothesis of the small B arising from B24 cannot be ruled out and is supported by the absence of the small B in samples of Zurita et al. (1998) and its appearance in a more recent sample (this paper).
Hypotheses for the origin of several B variants in Eyprepocnemis plorans. The 180 bp DNA repeat is shown in orange and the rDNA in green. (a) Simultaneous origin of B24 and the small B in the Torrox population by means of unequal crossing-over (double arrow) and subsequent deletion of rDNA in B24 or 180 bp DNA repeats in the small B. The star marks the B chromatid involved in these changes. An independent origin is also possible (dotted arrow) in which the small B arose from B24 by deletion of 180 bp DNA repeats and amplification of rDNA. (b) Origin of the rare large B variant found at Smir, by means of a paracentric inversion and subsequent amplification of rDNA in the interstitial band. Dotted arrows on the left show inversion break points.
The mechanism of origin for the large metacentric B found in the Salobreña population is centromere misdivision, as has been documented before (Henriques-Gil et al., 1984; López-León et al., 1993). This is justified by the symmetrical localization of both DNA probes with respect to the centromere. Finally, the large B found in a single male from Smir (Fig. 1c, lower row) probably arose by a paracentric inversion of the mid region involving about half of the rDNA and half of the 180 bp repeat of a B1 chromosome (Fig. 2b). The fact that this B is larger than B1 suggests that some DNA amplification has occurred after the inversion. Double FISH suggests amplification of the interstitial rDNA band (see Fig. 1c, lower row and Fig. 2b).
The principal B types (B1, B2 and B5) lack drive at present (López-León et al., 1992) and thus behave as near-neutral elements (Camacho et al., 1997a,b). The most plausible hypothesis suggests that they were selfish when they first invaded natural populations, but have been neutralized by the evolution of genes suppressing drive in the host genome (Herrera et al., 1996; Camacho et al., 1997b). Our model for the long-term evolution of parasitic B chromosomes states that a new parasitic B (showing drive) that emerged in a population containing a neutralized B (drive suppressed), could replace the neutralized B and regenerate the B polymorphism (Camacho et al., 1997a,b; Zurita et al., 1998; see also Camacho et al., 1999). It is clear that B24 illustrates this possibility, because this B type has recently replaced B2 in the only natural population where B24 has been found. The mean transmission ratio of B24 through females was about 0.7, which represents sufficient drive to explain its spectacular frequency increase in only 10 years, reaching the highest mean number of Bs per individual (1.53) ever found in a natural population of this species (Zurita et al., 1998).
The fact that B24 is the B-type showing the most extreme bias in favour of the 180 bp tandem repeat content (five times more than rDNA), suggests that B drive might depend on the relative amount of this repetitive DNA. As discussed above, B1 is considered the ancestral B type, but it has been replaced by B2 in the province of Granada and the eastern part of Málaga and by B5 in Fuengirola (Málaga). Likewise, B2 has recently been substituted by B24 in Torrox (Henriques-Gil & Arana, 1990; Camacho et al., 1997a,b; Zurita et al., 1998). Such substitutions constituted polymorphism regenerations by which a neutralized B lacking drive was replaced by a new selfish B showing drive (Camacho et al., 1997b). It is noteworthy that in all putative regenerations the new selfish B possessed relatively more 180 bp DNA repeat than the original neutralized B (e.g. B2 > B1, B5 > B1, B24 > B2). This suggests a possible role of this DNA sequence in B chromosome drive that should be investigated in the future.
References
Bakkali, M., Cabrero, J., López-León, M. D., Perfectti, F. and Camacho, J. P. M. (1999). The B chromosome polymorphism of the grasshopper Eyprepocnemis plorans in North Africa. I. B variants and frequency. Heredity. 83: 428–434.
Beukeboom, L. W. (1994). Bewildering Bs: an impression of the 1st B-Chromosome Conference. Heredity. 73: 328–336.
Cabrero, J., Alché, J. D. and Camacho, J. P. M. (1987). Effects of B chromosomes on the activity of nucleolar organizer regions in the grasshopper Eyprepocnemis plorans: activation of a latent nucleolar organizer region on a B chromosome fused to an autosome. Genome. 29: 116–121.
Camacho, J. P. M., Cabrero, J., Viseras, E., López-León, M. D., Navas-castillo, J. and Alché, J. D. (1991). G banding in two species of grasshopper and its relationship to C, N, and fluorescence banding techniques. Genome. 34: 638–643.
Camacho, J. P. M., Cabrero, J., López-León, M. D. and Shaw, M. W. (1997a). Evolution of a near-neutral B chromosome. In Henriques-Gil, N., Parker, J. S. and Puertas, M. J. (eds) Chromosomes Today, 12: 301–318. Chapman & Hall, London.
Camacho, J. P. M., Shaw, M. W., López-León, M. D., Pardo, M. C. and Cabrero, J. (1997b). Population dynamics of a selfish B chromosome neutralized by the standard genome in the grasshopper Eyprepocnemis plorans. Am Nat. 149: 1030–1050.
Camacho, J. P. M., Sharbel, T. F. and Beukeboom, L. W. (1999). B chromosome evolution. Phil Trans R Soc B. 354: in press.
Eickbush, D. G., Eickbush, T. H. and Werren, J. H. (1992). Molecular characterization of repetitive DNA sequences from a B chromosome. Chromosoma. 101: 575–583.
Gerlach, W. L. and Bedbrook, J. R. (1979). Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acids Res. 7: 1869–1885.
Henriques-Gil, N. and Arana, P. (1990). Origin and substitution of B chromosomes in the grasshopper Eyprepocnemis plorans. Evolution. 44: 747–753.
Henriques-Gil, N., Santos, J. L. and Arana, P. (1984). Evolution of a complex polymorphism in the grasshopper Eyprepocnemis plorans. Chromosoma. 89: 290–293.
Herrera, J. A., López-León, M. D., Cabrero, J., Shaw, M. W. and Camacho, J. P. M. (1996). Evidence for B chromosome drive suppression in the grasshopper Eyprepocnemis plorans. Heredity. 76: 633–639.
John, U. P., Leach, C. R. and Timmis, J. N. (1991). A sequence specific to B chromosome of Brachycome dichrosomatica. Genome. 34: 739–744.
Karpen, G. H. and Allshire, R. C. (1997). The case for epigenetic effects on centromere identity and function. Trends Genet. 13: 489–496.
López-León, M. D., Cabrero, J., Camacho, J. P. M., Cano, M. I. and Santos, J. L. (1992). A widespread B chromosome polymorphism maintained without apparent drive. Evolution. 46: 529–539.
López-León, M. D., Cabrero, J., Pardo, M. C., Viseras, E., Camacho, J. P. M. and Santos, J. L. (1993). Generating high variability of B chromosomes in the grasshopper Eyprepocnemis plorans. Heredity. 71: 352–362.
López-León, M. D., Neves, N., Schwarzacher, T., Heslop-Harrison, T. S., Hewitt, G. M. and Camacho, J. P. M. (1994). Possible origin of a B chromosome deduced from its DNA composition using double FISH technique. Chromosome Res. 2: 87–92.
López-León, M. D., Vázquez, P., Hewitt, G. M. and Camacho, J. P. M. (1995). Cloning and sequence analysis of an extremely homogeneous tandemly repeated DNA in the grasshopper Eyprepocnemis plorans. Heredity. 75: 370–375.
Zurita, S., Cabrero, J., López-León, M. D. and Camacho, J. P. M. (1998). Polymorphism regeneration for a neutralized selfish B chromosome. Evolution. 52: 274–277.
Acknowledgements
This study was supported by grants from the Spanish Dirección General de Enseñanza Superior (no. PB96–1433) and Plan Andaluz de Investigación, Grupo no. CVI-165 (Spain).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cabrero, J., López-León, M., Bakkali, M. et al. Common origin of B chromosome variants in the grasshopper Eyprepocnemis plorans. Heredity 83, 435–439 (1999). https://doi.org/10.1038/sj.hdy.6885960
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6885960
Keywords
This article is cited by
-
Out of patterns, the euchromatic B chromosome of the grasshopper Abracris flavolineata is not enriched in high-copy repeats
Heredity (2021)
-
Transcription of a B chromosome CAP-G pseudogene does not influence normal Condensin Complex genes in a grasshopper
Scientific Reports (2017)
-
Next generation sequencing and FISH reveal uneven and nonrandom microsatellite distribution in two grasshopper genomes
Chromosoma (2015)
-
B chromosomes showing active ribosomal RNA genes contribute insignificant amounts of rRNA in the grasshopper Eyprepocnemis plorans
Molecular Genetics and Genomics (2014)
-
HP1 knockdown is associated with abnormal condensation of almost all chromatin types in a grasshopper (Eyprepocnemis plorans)
Chromosome Research (2014)