Abstract
In northern Greece, insecticides have been used intensively against aphid populations of the Myzus persicae group, both on its primary host peach, on which annual sexual reproduction occurs, and on secondary host field crops such as tobacco, on which reproduction is entirely parthenogenetic. This has resulted in the selection of high levels of resistance based on the amplification of two genes encoding insecticide-degrading esterases, E4 and FE4. We have used fluorescence in situ hybridization (FISH) to study variation in the number and distribution of loci with amplified esterase genes in clones established from field populations of M. persicae- group aphids sampled on various crops in northern Greece. All clones collected from peach, as well as most of the clones (74%) from tobacco and other secondary host plants, had amplified FE4 genes and were of normal karyotype. Amplicon clusters containing FE4 occurred at multiple sites which varied in number, zygosity and distribution between clones. Most loci were on autosome 1, which also had the only site that was consistently occupied by amplified FE4, situated near subtelomeric repetitive DNA. Possibly this was the original site of FE4 gene amplification, and the location of the single-copy ‘wild type’ esterase genes. The rest of the clones from tobacco and other secondary hosts (26%) had amplified E4 genes, and all those analysed by FISH had an amplicon cluster on autosome 3T at a site close to the breakpoint of an A1,3 translocation, confirming the close linkage of this translocation with, and its probable involvement in, E4-based resistance. Three translocated clones collected on capsicum at one site had both E4 and FE4 amplified, the first time that both esterase gene types have been found together in individual aphids from field populations.
Similar content being viewed by others
Introduction
The economically important pest aphid Myzus persicae (Sulzer) can combat organophosphate and carbamate insecticides by overproducing insecticide-degrading esterases, encoded by amplified genes (Devonshire & Field, 1991). Resistant aphids have one of two different but closely related amplified genes, E4 or FE4, according to their karyotype. Aphids heterozygous for an autosomal (A) 1,3 translocation have amplified E4, whereas those without this translocation have FE4 (Blackman et al., 1995). Studies of the inheritance of these genes, coupled with in situ hybridization to map their locations on chromosomes (Blackman et al., 1996), have shown that E4- containing amplicon clusters are usually located at one site on autosome 3T closely linked to the A 1,3 translocation, whereas in clones with FE4-based resistance the amplicon clusters occurred at three to five sites on several different chromosomes.
Myzus persicae has a complex pattern of life cycle variation (Blackman, 1974). Populations on field crops reproduce by thelytokous (all-female), apomictic parthenogenesis, but in temperate regions a variable proportion of the population migrates in autumn to peach trees (Prunus persica), where an annual bisexual generation occurs with meiotic recombination and overwintering of cold-resistant eggs. The proportion migrating to peach trees varies geographically according to (i) seasonal trends in photoperiod and temperature and (ii) the availability of peach. The karyotype (presence or absence of the A 1,3 translocation) and the type of esterase produced (E4 or FE4), appears to be associated with the life cycle category, in a way that is relevant to the evolution and inheritance of insecticide resistance. Translocated, E4-producing genotypes are widely distributed in warm temperate (and tropical) regions of the world, and in glasshouses in northern temperate regions. Mild winters allow such populations to reproduce all the year around by parthenogenesis, and most of them have lost the bisexual part of the life cycle. On the other hand, FE4-based resistance seems to have developed in peach-growing areas, in the absence of the translocation, where at least part of the population goes through the bisexual phase.
In some places, such as northern Greece, climate and availability of peaches favour both life cycle categories to a similar extent, and intensive use of insecticides in peach orchards and on field crops has resulted in high levels of resistance, so that genotypes with FE4-based resistance occur on peach trees in spring, and genotypes with either E4- or FE4-based resistance occur together in field crops. Myzus persicae-group populations in northern Greece also show much genetic variation in other respects, such as body colour and adaptation to different host plants; tobacco is widely grown, and populations on tobacco are sufficiently distinct to be recognized as a separate taxon (nicotianae; Blackman & Spence, 1992). Northern Greece therefore seemed to be an ideal place to examine the extent of variation in number and distribution of loci with amplified esterase genes that can occur in field populations of M. persicae-group aphids.
Materials and methods
Aphid collection and rearing
We sampled populations on peaches and some crop plants in northern and central mainland Greece in early May 1995, and populations on field crops in late May — mid April 1996 (Table 1). Clones were set up using the progeny of single parthenogenetic females collected from widely separated plants at each location. They were maintained at the Natural History Museum on excised leaves of potato (var. Pentland Crown) in a controlled environment room at 16 h photoperiod and 15°C.
Estimation of carboxylesterase activity
Assays for carboxylesterase activity were carried out on homogenates of individual aphids from each of the 1995 clones. The amount of detoxifying enzyme present was measured in a small fraction (one-fiftieth) of each homogenate by immunoassay using E4 antiserum in 96-well microtitre plates (Devonshire et al., 1992). Three individuals of each clone were categorized as either susceptible (S) or resistant (R1, R2 or R3, depending on the amount of enzyme present compared with aphids from standard laboratory clones). Immunoassays were also performed on 1996 clones, but the data had to be discounted because of major inconsistencies between replicates.
Esterase gene characterization, karyotyping and FISH
The type(s) of esterase gene amplified in each clone (if any) was identified by a PCR-based diagnostic (Field et al., 1996). In some key clones, especially those which had both E4 and FE4, this was confirmed by probing Southern blots of genomic DNA digested with MspI (Blackman et al., 1996). Presence or absence of the A 1,3 translocation was observed in preparations of embryonic somatic cells viewed by phase contrast, which were then hybridized in situ to a biotinylated 8 kb genomic DNA probe including most of the E4 gene and some 3§ flanking DNA. Under the conditions used, this probe hybridized to both E4 and FE4 sequences. Methods of chromosome preparation and fluorescence in situ hybridization have already been described (Blackman et al., 1995).
Chromosome mapping
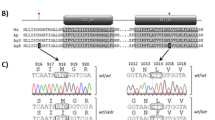
The chromosomes of Hemiptera are holocentric, so lack physical points of reference, and there have been no classical genetic studies of M. persicae (or any other aphid species) to provide linkage maps. In order to describe the locations of the amplified esterase sequences, we have divided the M. persicae haploid karyotype (n=6) into 42 sections, and labelled the autosomal sections consecutively as A–Z on the longer autosomes 1–3, and a–f on the short autosomes 4 and 5 (Fig. 1a). The terminal sections of each autosome are defined as comprising telomeric and subtelomeric DNA; the latter largely consists of numerous copies of a 169 bp repeat (MpR: Spence et al., 1998). After observation of a number of cells (usually six to 10) of each clone it is usually possible to assign the locations of the hybridized probe to specific chromosome segments. In order to apply this labelling scheme to the A1,3 translocation karyotype, we defined this translocation as involving breaks within section I of autosome 1 and section W of autosome 3, so that the subterminal section of autosome 1 was exchanged for sections S–V plus W (part) of autosome 3 (Fig. 1b, cf. Fig. 1a). This is supported by recent work indicating that the subtelomeric repetitive sequence is relocated from autosome 1 onto autosome 3T in A 1,3 translocation heterozygotes of M. persicae (Spence et al., 1998).
Results
All 35 clones established from the 1995 collections from peach in early May were of normal karyotype (i.e. none had the A 1,3 translocation), and all had amplified FE4 genes. FISH revealed that FE4 amplicon clusters occurred at six different loci (at least) on autosome 1 (although the maximum number discernible in any one clone was four), at two loci on autosome 3, and at one locus near the end of a short autosome, probably autosome 5 (Table 2 and Fig. 2a,b,c,d,e and f). The last site was only found in one clone (F2), which had a total of eight sites with amplified sequences at six loci, the largest number of amplicon clusters so far recorded (Fig. 2a). Up to three of the loci on autosome 1 were homozygous for amplified FE4 in any one clone (Fig. 2b). All six of the 1995 clones heterozygous for amplified FE4 in segment W on autosome 3 were from the same peach orchard (at Falani), but these six clones differed with respect to the other loci amplified. Clones started from aphids collected on tobacco and sugar beet plants in fields adjacent to the sampled peach orchards had a range of distribution and number of FE4 amplicon clusters similar to those from peach trees.
In situ hybridization of an esterase (E4) DNA probe to chromosomes of insecticide-resistant Myzus persicae from Greece, showing location of amplified esterase genes. FE4 amplicon clusters on autosome 1 are indicated by small thin arrows, those on other autosomes by arrowheads: (a) clone F2, with eight amplicon clusters, including sites on autosomes 3 and 5; (b) clone B3, with six amplicon clusters on autosome 1, interpreted as three homozygous loci; (c) clone F1, with five amplicon clusters, interpreted as two homozygous loci on autosome 1 and an additional site on autosome 3; (d) clone E2, with six amplicon clusters on autosome 1, interpreted as two homozygous loci plus additional sites in segment D of one homologue and segment E of the other; (e) clone B1, with five amplicon clusters on autosome 1 at three loci — the most frequent combination of sites found in the populations studied; (f) clone 9656, which has the autosome 1,3 translocation found in genotypes with amplified E4, and also has amplified FE4. From previous work it is probable that the two amplicon clusters on one homologue of autosome 1 are FE4, and that the cluster at one end of the short element (3T) is E4. Bar represents 10 μm.
Immunoassays for carboxylesterase activity showed that all the 1995 clones, with from two to eight amplicon clusters, had moderate to high (R2 or R3) levels of insecticide resistance.
Of the 184 clones set up from 1996 collections on field crops, 46 (26%) had the A1,3 chromosome translocation. Most of these (78%) were from the two tobacco fields sampled. All of the translocated clones had amplified E4, but three clones originating from capsicum (9636, 9651 and 9656) also had amplified FE4. Clones with both amplified genes have been obtained in laboratory crosses (Blackman et al., 1996), but this is the first time that they have been found together in the field. The body colour of the three clones with both E4 and FE4 was green, whereas all other clones with the A1,3 translocation were red. The clones of normal (untranslocated) karyotype included both red and green colour morphs, with green predominating (82%), but all had only FE4 amplified, irrespective of body colour. There was no correlation between colour and number of FE4 amplicon clusters.
FISH to 22 of the 1996 clones did not reveal any amplicon clusters that were additional to those seen in the 1995 clones (Tables 1 and 2). Two of the clones from capsicum were apparently homozygous for amplified FE4 at the W locus on autosome 3. The three clones from capsicum with both E4 and FE4 amplified had two sites (presumably FE4) on one homologue of autosome 1, plus the presumed E4 site on autosome 3T (Fig. 2f). Three translocated clones with only E4 amplified, originating from three different crops, all had a single amplicon cluster on autosome 3T (Table 2), as found previously in other E4-producing, translocated clones from the U.K. and the U.S.A. (Blackman et al., 1995).
In FE4-producing clones, the only locus that was invariably amplified on at least one homologue was that in segment 1I, the site closest to the end of autosome 1 (Table 2).
Discussion
It is only possible to map the approximate positions of the amplified FE4 genes so, if anything, the number of separate locations may be underestimated. However, one can have some confidence in the single identity of the most commonly occupied sites (1D,1H, 1I, 3W), as one would expect these to become homozygous for amplified FE4 in sexually reproducing populations. The patterns of presumed homozygosity/heterozygosity at these sites (Table 2) conform to this expectation.
Our FISH technique was not sufficiently sensitive to locate the single-copy ‘wild-type’ FE4 locus in susceptible aphids, so we do not know whether amplification has occurred at the original locus. In Chinese Hamster ovary (CHO) cell lines, initial amplification of dihydrofolate reductase (DHFR) genes occurs on the same chromosome as the parental single-copy gene but usually at some distance (at least 50 Mb) away from it (Trask & Hamlin, 1989). In M. persicae, in spite of all the variation among clones in the number and distribution of FE4 amplicon clusters, there is only one site (1I) where they occur consistently on at least one homologue, suggesting that this is the original site of the amplification, and possibly of the single-copy parental FE4 and E4 genes, which are thought to occur adjacent to each other in a head-to-tail arrangement, with E4 upstream of FE4 and ≈19 kb of intervening sequence (Field & Devonshire, 1998).
Contrary to our initial conclusions based on studies of laboratory clones (Blackman et al., 1995), it now seems that, at least in Greek populations of M. persicae, other locations of FE4 amplicon clusters occur most commonly on the same chromosome (autosome 1) as the putative original amplification site, similar to the situation found in CHO cells. Subsequent relocation of FE4-containing amplicons to other sites on autosome 1 may have occurred by inversion, and to other autosomes by reciprocal interchange. The postulated original locus (1I) is situated near subtelomeric heterochromatin containing numerous copies of the 169 bp repeat MpR that also occurs subtelomerically on all the other autosomes of M. persicae (Spence et al., 1998), suggesting that interchange of subtelomeric sections may occur relatively frequently.
The MpR subtelomeric repeat has been shown to have relocated from autosome 1 onto autosome 3T in clones with the A 1,3 translocation. An interchange event with similar breakpoints, followed by recombination in a subsequent sexual phase, could have led to the occurrence of FE4 amplicon clusters at site 3W. Transposable elements might be involved in any or all of such rearrangements. In this context, a non-LTR retrotransposon-like sequence associated with the telomeric DNA of M. persicae has recently been isolated (Spence, unpublished results), and inverted repeat sequences have been found within the E4 amplicon (Field, unpublished data).
If FE4 gene amplification was a unique event, then this most likely happened in M. persicae populations on peaches in north-western U.S.A., where organophosphate resistance was first reported in the mid-1950s (Anthon, 1955). This was more than 10 years before the first reports of resistance in peach-feeding populations in some parts of the Mediterranean area (Baranyovits, 1973). However, such resistance was not noted in peach- and tobacco-growing areas of northern mainland Greece until about 1985, and in 1989 it still seemed to be limited to two localities (G. Michalopoulos, personal communication). The large variation in number and distribution of FE4-containing amplicon clusters now present may therefore be a rather recent development. In contrast to results with clones of known parentage reared in the laboratory (Blackman et al., 1996), no correlation was found in 1995 Greek field-collected clones between the number of amplicon clusters and carboxylesterase activity, suggesting that FE4 gene copy number may be less variable than the number of sites at which it is located. It will be of interest to examine populations with FE4-based resistance in other countries and other parts of the world, to see whether different patterns of amplicon cluster distribution occur outside Greece, indicating that multiple loci have arisen independently. That this may be the case is suggested by one clone from France used in studies of the inheritance of resistance (French R; Blackman et al., 1995); this had an amplicon cluster at the 1I site as usual, but was also homozygous for amplified FE4 at a subtelomeric locus on autosome 2, a site not occupied by amplified FE4 genes in any of the Greek clones studied.
References
Anthon, E. W. (1955). Evidence for green peach aphid resistance to organophosphorus insecticides. J Econ Entomol, 48: 56–57.
Baranyovits, F. (1973). The increasing problem of aphids in agriculture and horticulture. Outlook on Agric, 7: 102–108.
Blackman, R. L. (1974). Life-cycle variation of Myzus persicae (Sulz.) (Hom., Aphididae) in different parts of the world, in relation to genotype and environment. Bull ent Res, 63: 595–607.
Blackman, R. L. and Spence, J. M. (1992). Electrophoretic distinction between the peach-potato aphid Myzus persicae and the tobacco aphid M. nicotianae (Homoptera: Aphididae). Bull ent Res, 82: 161–165.
Blackman, R. L., Spence, J. M., Field, L. M. and Devonshire, A. L. (1995). Chromosomal location of the amplified esterase genes conferring resistance to insecticides in the aphid Myzus persicae. Heredity, 74: 297–302.
Blackman, R. L., Spence, J. M., Field, L. M., Javed, N., Devine, G. and Devonshire, A. L. (1996). Inheritance of the amplified esterase genes responsible for insecticide resistance in Myzus persicae (Homoptera: Aphididae). Heredity, 77: 154–167.
Devonshire, A. L. and Field, L. M. (1991). Gene amplification and insecticide resistance. Ann Rev Ent, 36: 1–23.
Devonshire, A. L., Devine, G. J. and Moores, G. D. (1992). Comparison of microplate esterase assays and immunoassay for identifying insecticide resistant variants of Myzus persicae (Homoptera: Aphididae). Bull ent Res, 82: 459–463.
Field, L. M. and Devonshire, A. L. (1998). Evidence that the E4 and FE4 genes responsible for insecticide resistance in the aphid Myzus persicae (Sulzer) are part of a gene family. Biochem J, 330: 169–173.
Field, L. M., Crick, S. E. and Devonshire, A. L. (1996). Polymerase chain reaction-based identification of insecticide resistance genes and DNA methylation in the aphid Myzus persicae (Sulzer). Insect Mol Biol, 5: 197–202.
Spence, J. M., Blackman, R. L., Testa, J. M. and Ready, P. D. (1998). A 169bp tandem-repeat DNA marker for sub-telomeric heterochromatin and chromosomal rearrangements in aphids of the Myzus persicae group. Chromosome Res, 6: 167–175.
Trask, B. J. and Hamlin, J. L. (1989). Early dihydrofolate reductase gene amplification events in CHO cells usually occur on the same chromosome arm as the original locus. Genes Devel, 3: 1913–1925.
Acknowledgements
This work was supported by a Linked Research Groups grant from the British Biotechnology and Biological Sciences Research Council. We thank Amanda Anderson, Emma De Boise, Zöe Harling and Diana Johnston for technical assistance; and George Kokkinis, George Skoulakis, Nikos Katis, Liliana Dobri, Philippos Ioannidis, John Margaritopoulos, John Tsitsipis and Costas Zarpas for assistance with location and collection of aphids in Greece. The aphids were brought to the U.K. from Greece under European Community Directive 95/44/EC and U.K. Ministry of Agriculture, Fisheries and Food Plant Health Licence no. PHF 1714 A,/1915.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Blackman, R., Spence, J., Field, L. et al. Variation in the chromosomal distribution of amplified esterase (FE4) genes in Greek field populations of Myzus persicae (Sulzer). Heredity 82, 180–186 (1999). https://doi.org/10.1038/sj.hdy.6884580
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6884580