Abstract
Seeds of conifers contain a female megagametophyte (endosperm), which is usually haploid. Results of biochemical analysis indicate the existence of a nonembryonic tissue in the seeds of Cupressus sempervirens, which expresses at isozyme gene loci the phenotype of the seed tree and is diploid. This paper presents the genetic verification of the existence of this tissue and discusses its possible origin.
Similar content being viewed by others
Introduction
Seeds of conifers contain two enzymatically active tissues: the female megagametophyte, also known as the seed endosperm, and the seed embryo. The megagametophyte is haploid and contains the nuclear genetic information of the female gamete. The embryo, or zygote, is in most cases diploid. The megagametophyte surrounds the embryo and is a nutritive tissue.
The megagametophyte has frequently been used in isoenzymatic studies of coniferous species. Studies of a sample of megagametophytes derived from a single tree provide information for segregation and linkage analysis. Another application is the identification of the genotype of the seed parent through the electrophoretic analysis of at least six megagametophytes giving a 95% probability of correct assignment (Morris & Spieth, 1978). Finally, the male contribution to the zygote can be inferred at single gene loci by a comparison of the genetic information contained in embryo and megagametophyte tissue (Müller, 1976).
Raddi et al. (1990) and later Papageorgiou et al. (1993) failed to isolate the megagametophyte from the seeds of the Mediterranean cypress (Cupressus sempervirens: Cupressaceae). The embryo could be separated easily, but the remaining tissue yielded zymograms which were characteristic of diploid tissue. This observation was repeated with different enzyme systems. As these zymograms were not of embryonic origin, another diploid tissue must have been responsible for these results. In the present work, the genetic verification of the existence of this tissue will be presented, and its possible origin will be discussed together with its application in genetic studies.
Materials and methods
Seeds were collected from various adult Mediterranean cypress trees (Cupressus sempervirens) growing in 23 populations in Greece: 18 natural stands (Crete, Samos, Kos, Rodos and Symi), two ornamental plantations (Athens) and three naturally expanding afforestations (Peloponnese and Kefallonia). Seeds were collected from an average of 20 single trees per provenance and carefully kept separately. Additionally, needles from 20 trees, growing in Athens, were collected.
Seeds of Cupressus sempervirens were also collected from two seed orchards in Tatoi (Athens) and Megalopoli (Peloponnese). Two grafted ramets (one from each seed orchard) from 111 clones were selected. These clones derived from original material representing four natural stands (Samos, Kos, Rodos and Symi) and two afforestations (Peloponnese). Seeds were collected from single trees and kept separately.
Seeds of C. lusitanica, C. macrocarpa and C. arizonica were also sampled from nine single seed trees (three of each species) growing in the seed orchards described above. As before, seeds of each single tree were kept separately.
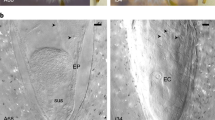
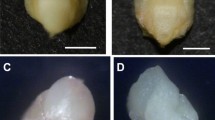
Samples of seeds were analysed using horizontal starch gel electrophoresis. In each seed, two tissues were separated, after the seed cover had been removed: the embryonic tissue, which was white in colour and was found in the centre of the seed; and the nonembryonic tissue, which surrounded the embryonic tissue. It was larger than the embryonic tissue and was whitish-yellow on the inside (next to the embryo) and brownish-yellow on the outside (next to the seed cover).
The two tissues were separated from each other easily. A diagram presenting the seed anatomy of C. sempervirens is given in Fig. 1.
Both tissues were analysed by electrophoresis in the laboratory for each seed, in order to compare their zymograms. The analysis was based on the following eight enzyme systems: two monomeric (Lap, Pgm), four dimeric (Pgi, 6-Pgdh, Ndh, Mdh) and two tetrameric (Gdh, Mnr) (see Table 1). Details of laboratory techniques were described by Bergmann (1974), Cheliak & Pitel (1984) and Wendel & Weeden (1989).
The results of an inheritance analysis of isoenzyme variation in C. sempervirens (Papageorgiou, 1995) were used to verify the results of the present study. Because there was no way to analyse megagametophytes, a method suitable for angiosperms was performed (Gillet & Hattemer, 1989). This involved formulating and testing a hypothesis for mode of inheritance based on the comparison between the zymogram of the seed parent and the patterns observed from its open-pollinated progeny. Under this hypothesis, each phenotype could be tracked back to only one genotype which was thus identifiable. For each seed parent with the genotype AiAj (i≠j), the following qualitative and quantitative relationships were expected to hold among its offspring from open-pollination:
(i) each offspring must possess at least either allele Ai or Aj,
(ii) Nij=Nii+Njj,
(iii) Nik=Njk (k≠i,j),
where Nxy denotes the number of offspring that carry the phenotype AxAy. Relationships (ii) and (iii) were quantitatively tested by the exact binomial goodness-of-fit test.
Results
The zymograms obtained from the analysis of embryonic and nonembryonic tissues of the seeds of Cupressus sempervirens were compared to each other and gave the following results.
(i) Comparisons between zymograms of embryonic and nonembryonic tissue were made for 30 seeds of the same seed tree. All nonembryonic zymograms were identical for all seeds belonging to the same tree and had a typical diploid pattern for all enzyme systems. The embryos showed different patterns and at all loci studied had at least one band in common with the nonembryonic zymogram.
(ii) Analysis of nonembryonic tissues of seeds of different trees (30 seeds from each tree) always showed no segregation among the seeds of a tree but large variation in patterns among trees, at all loci studied.
(iii) Zymograms of nonembryonic tissue obtained from seeds of different ramets (30 seeds from each ramet) belonging to the same clone always showed identical zymograms at all loci studied.
(iv) In order to determine whether the nonembryonic tissue was of sporophytic origin, the zymograms of nonembryonic tissues of 20 trees were compared with the zymograms of needles sampled from the same trees. At all gene loci examined, all nonembryonic tissues showed zymograms identical to those of the needles.
(v) In order to determine the mode of inheritance of the zymograms, inheritance analysis of isoenzyme variation in C. sempervirens was performed by the method of Papageorgiou (1995). The nonembryonic tissue was considered to express the seed parent. The proportions of the progeny zymograms were not significantly different at any of the gene loci examined from those expected under regular gametic segregation and random fusion. Thus the hypothesis was not rejected for any of the gene loci. The results of the inheritance analysis are presented in Table 2
The results obtained indicate the existence of a nonembryonic tissue in the seeds of C. sempervirens, which expresses the phenotype of the seed tree at isozyme gene loci. Figure. 2 presents an electrophoresis gel scored for the dimeric enzyme system Pgi. The locus identified is called Pgi-B. The nonembryonic tissue of the seed tree U28 (Crete) was compared with 17 embryos from seeds of the same tree. All embryonic tissues were found to possess at least one allele in common with the nonembryonic tissue (B1B2) and the proportion of genotypes between the embryonic tissues was not significantly different from that expected under the hypothesis of a codominant single locus: N12=8; N11+N22=9 (Nxy denotes the number of embryos that carry the genotype BxBy).
Inheritance analysis of Cupressus sempervirens at Pgi-B. The leftmost zymogram expresses the genotype of the parent tree (nonembryonic tissue), and the remaining 17 express the genotype of its progeny (embryos). Interpretation (from left to right): seed tree genotype: B1B2; progeny genotypes: B2B2, B1B2, B1B2, B1B1, B2B2, B2B2, B2B2, B2B2,B1B2, B1B2, B1B2, B2B2, B1B2,B1B2,B1B2,B2B2, B2B2.
This phenomenon has not yet been reported in any other species, not even in members of the genus Cupressus, such as C. macrocarpa. (Kafton, 1976; Conkle, 1986). In order to verify this result, an isoenzyme analysis was performed for seeds of trees belonging to three species of the genus Cupressus: C. lusitanica, C. macrocarpa and C. arizonica. The zymograms of the embryonic and the nonembryonic tissues of each seed were compared, as for C. sempervirens. Fourteen seeds were analysed for each tree. Zymograms of nonembryonic tissues of the same seed tree were segregating for all three species: C. lusitanica at Lap, C. macrocarpa at 6-Pgdh and C. arizonica at Pgi. This follows the general rule, which is valid for all conifers, but apparently not for C. sempervirens.
Discussion
The development of the megagametophyte in Mediterranean cypress does not seem to differ from that described for other conifers (Singh, 1978). The expansion of the megagametophyte through mitotic divisions of the haploid gamete causes degeneration of the diploid sporophytic nucellus. In the stage of a fully developed seed, the megagametophyte is large and surrounds the embryo. A brown, degenerated layer surrounds the megagametophyte, which represents the remainder of the nucellus. According to Singh (1978) this layer is strongly pressed, thin and fleshy.
The genetic results have demonstrated the existence of a diploid sporophytic tissue in the nonembryonic part of the Mediterranean cypress seed. Raddi et al. (1990) have proposed the term ‘seed perisperm’ to describe the ‘diploid maternal tissue of the seed’. Papageorgiou et al. (1993) have noted the possibility that some cells of the diploid nucellus might have been active and not degenerated completely. However, a protein analysis (S. Raddi, pers. comm.) did not show any activity in this particular tissue. Pichot & El Maataoui (1997) have used flow cytometry to test the ploidy level of several coniferous seeds and have confirmed the observation of the previous studies. They suggested multiple ploidy levels for some members of the family Cupressaceae. Cupressus sempervirens and C. arizonica were among them. Yet these results cannot explain the existence of two different alleles in the nonembryonic tissue of a heterozygous seed tree nor why this happens especially in C. sempervirens and not in C. arizonica or C. lusitanica.
The present study has proved the existence of sporophytic diploid tissue in the seed of C. sempervirens. The origin of this tissue, and why it occurs only in the Mediterranean cypress, is not known at this stage. Further cytological studies are needed to give an accurate explanation.
This diploid tissue has already been used to identify the genotypes of seed parents for clone identification (Raddi et al., 1990), inheritance analysis (Papageorgiou et al., 1993; Papageorgiou, 1995) and population studies (Papageorgiou et al., 1994). It is much more efficient than the megagametophyte, because each seed, no matter how far it is found from its seed parent, gives its exact multilocus genotype. This reduces the sample size needed for seed tree identification and can become a unique tool for studies of seed dispersal.
References
Bergmann, F. (1974). The genetics of some isoenzyme systems in spruce endosperm (Picea abies). Genetica, 6: 353–360.
Cheliak, W. M. and Pitel, J. A. (1984). Techniques for Starch Gel Electrophoresis of Enzymes from Forest Tree Species. Information Report PI-X-42. Petawawa National Forestry Institute, Canadian Forestry Service, Chalk River, Ontario.
Conkle, M. T. (1986). Electrophoretic analysis of variation in native Monterey cypress (Cupressus macrocarpa Hartw.). In: Elias, T. S. (ed.) Proceedings of a California Conference on the Conservation and Management of Rare and Endangered Plants, pp. 249–256. The California Native Plant Society, Sacramento, CA.
Gillet, E. and Hattemer, H. H. (1989). Genetic analysis of isoenzyme phenotypes using single tree progenies. Heredity, 63: 135–141.
Kafton, D. L. (1976).Isozyme Variability and Reproductive Phenology of Monterey Cypress. Ph.D. Thesis, University of California, Berkeley, CA.
Morris, R. W. and Spieth, P. T. (1978). Sampling strategies for using female gametophytes to estimate heterozygosity in conifers. Theor Appl Genet, 51: 217–222.
Müller[-Starck], G. (1976). A simple method of estimating rates of self-fertilization by analysing isozymes in tree seeds. Silvae Genet, 25: 15–17.
Papageorgiou, A. C. (1995). Genetische Untersuchungen zur Züchtung und Generhaltung der Mittelmeerzypresse (Cupressus sempervirens L.) in Griechenland. Göttingen Res Notes Forest Genet, 18: 1–190.
Papageorgiou, A. C., Bergmann, F., Gillet, E. and Hattemer, H. H. (1993). Genetic analysis of isoenzyme variation in Mediterranean cypress (Cupressus sempervirens L.). Silvae Genet, 42: 109–111.
Papageorgiou, A. C., Panetsos, K. P. and Hattemer, H. H. (1994). Genetic differentiation of natural Mediterranean cypress (Cupressus sempervirens L.) populations in Greece. Forest Genet, 1: 1–12.
Pichot, C. and Elmaataoui, M. (1997). Flow cytometric evidence for multiple ploidy levels in the endosperm of some gymnosperm species. Theor Appl Genet, 94: 865–870.
Raddi, S., Di Lonardo, V. and Sufra, M. (1990). The use of biochemical markers to distinguish cypress clones. In:Progress in EEC Research on Cypress Diseases pp. 43–49. Agrimed Research Programme. Commission of the European Communities, Agriculture, Report EUR 12493 EN.
Singh, S. (1978). Embryology of Gymnosperms. Encyclopedia of Plant Anatomy. Gebrüder Borntraeger, Berlin.
Wendel, J. F. and Weeden, N. F. (1989). Visualization and interpretation of plant isozymes. In: Soltis, D. E. and Soltis P. S. (eds) Isozymes in Plant Biology. pp. 5–45. Chapman and Hall, London.
Acknowledgements
The author wishes to thank F. Aravanopoulos, F. Bergmann, S. Fileri, R. Finkeldey, E. Gillet, H.-R. Gregorius, H. H. Hattemer, L. Leinemann, M. Kafka, S. Raddi and E. Teissier du Cros for helpful discussions. This study was financed by the European Community (DG VI).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Papageorgiou, A. Diploid sporophytic tissue in the seed of Cupressus sempervirens L.. Heredity 81, 586–590 (1998). https://doi.org/10.1046/j.1365-2540.1998.00430.x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00430.x
Keywords
This article is cited by
-
Genetic differences between autochthonous and breeding populations of common cypress (Cupressus sempervirens L.) in Greece
European Journal of Forest Research (2005)