Abstract
Genetic variability of 22 populations of Helicoverpa armigera (Hübner) (Lepidoptera, Noctuidae) was studied by comparison of allozyme frequencies at 15 loci. Seven loci were polymorphic at the 99% level. Populations originated from the south of France, Portugal, Morocco, Tunisia, Burkina Faso and Ivory Coast. Few significant deviations from Hardy–Weinberg equilibrium occurred, except for the Acp locus. Overall comparison of allele frequencies revealed significant differences for four loci, although the value of θ was low (0.007). At a lower geographical scale, two situations were observed. Populations from France and Portugal showed significant differences when comparing allele frequencies, although θ was low (0.0099). On the other hand, in northern and western Africa, no significant difference exists even when comparing populations located on either side of the Sahara desert. Results are discussed in relation to the migratory abilities of the species.
Similar content being viewed by others
Introduction
Helicoverpa armigera (Hübner) is a cosmopolitan pest, whose distribution area covers all the Old World, approximately between the northern and southern fortieth parallels (Hardwick, 1965). In Burkina Faso, H. armigera breeds in two types of asynchronous agrosystems (Nibouche, 1994). During the rainy season, from mid-June to October, the pest colonizes rainfed crops (mainly cotton and maize) and weeds. Throughout the dry season, from October to mid-April, H. armigera attacks irrigated crops. During the 2-month period between mid-April and mid-June, irrigated crops are harvested and no population of H. armigera is noticed. Helicoverpa armigera has both migratory and diapausing abilities that allow colonization of ephemeral habitats and survival in unfavourable conditions (Cayrol et al., 1974). Biological studies have shown that diapause occurs in Burkina Faso, but at very low rates (Nibouche, 1994). Seasonal migrations could occur between rainfed crops and irrigated crops within Burkina Faso or, on a greater scale, between the Sudanese climatic area (Burkina Faso) and the Guinean climatic area (Ivory Coast). Such migrations following the seasonal movements of the intertropical convergence zone have been documented in Heteroptera of the genus Dysdercus (Duviard, 1981) and in Agrius convolvuli L. (Lepidoptera, Sphingidae) (Bowden, 1973).
Despite its limits (Hillis & Moritz, 1990), the study of allozyme polymorphism is well suited to population studies and has been used to study the genetic structure of populations in several migrant Noctuidae (Daly & Gregg, 1985; Pashley et al., 1985; Korman et al., 1993; Buès et al., 1994). In order to assess the extent of migration of H. armigera in western Africa, we have studied allozyme polymorphisms in populations from Burkina Faso and Ivory Coast. Populations from northern Africa and southern Europe were included in this study to control for a possible lack of genetic structure by comparing the amount of gene flow that occurs in western Africa with that encountered over a larger area.
Materials and methods
Collections
Samples originated from 22 locations in Burkina Faso, Ivory Coast, France, Morocco, Portugal and Tunisia (Table 1) and Fig. 1). Locations less than 50 km distant from one another were grouped in ‘regions’ (south-east France, Ouagadougou and Bobo-Dioulasso). Three ‘zones’ were defined (Fig. 1): France and Portugal, northern Africa (Morocco and Tunisia) and western Africa (Burkina Faso and Ivory Coast).
Except for the sample from Plateau de Lhers (moths collected with a light trap), populations were sampled as larvae from several host plants (Table 1). Larvae were reared on an artificial diet (Poitout & Buès, 1974; Giret & Couilloud, 1986) until pupation. After emergence, adults were frozen at −80°C.
Electrophoresis
Moths were separated into abdomen and thorax plus head samples. The thorax plus head samples were homogenized in 120 μL of a Tris–borate–EDTA buffer (pH 7.0). Horizontal starch gel electrophoresis was carried out according to a method adapted from Pasteur et al. (1987). Gel and electrode buffer was a Tris–maleate buffer (pH 7.3). Fifteen enzymes were studied (Table 2). Enzyme staining recipes were adapted from Pasteur et al. (1987) and from Hillis & Moritz (1990).
Data analysis
Mean heterozygosity (Pasteur et al., 1987) and mean genetic distance (Nei, 1978) of the 22 populations for the 15 studied loci were calculated.
Deviation from Hardy–Weinberg equilibrium was tested using two methods. The first consisted of calculating F (= FIT, which measures deviations of the whole population) and f (= FIS, which measures deviations within populations) according to the Weir & Cockerham (1984) formulae, using the computer program DIPLOID (Weir, 1990). Standard deviations were calculated by the jackknife method. The second method compared the observed genotypical proportions with expected Hardy–Weinberg proportions using the exact Fisher test computed by GENEPOP (Raymond & Rousset, 1995).
Three methods were used to assess the genetic structure of populations: (i) comparison of allele frequencies; (ii) calculation of the θ index (Weir & Cockerham, 1984), which is a weighted calculation of Wright's (1951) FST that measures genetic difference between populations; and (iii) the private allele method of Slatkin (1985).
Allele frequencies were compared within each locus using the Fisher test (computed by GENEPOP). A global χ2-test across loci was made by the Fisher method (Fisher, 1938). To allow this global χ2-test, loci were considered as independent. Calculations were performed at several geographical scales (within zones or regions, between all populations) to define groups of homogeneous populations exhibiting no significant genetic variation. Comparisons were also made after pooling of the populations to increase the power of tests. Pooling was carried out only when there was no significant difference within the considered area.
DIPLOID allowed the calculation of θ with a standard deviation computed by jackknife across loci. The value of Nm, the mean number of efficient migrants exchanged between populations at each generation, was estimated from θ within the Wright's (1931) island model, in which the global population is subdivided into subpopulations. A value of Nm≲1 is considered sufficient to make gene flow overcome genetic drift (Wright, 1931; Spieth, 1974). Nm was estimated from θ as follows (Wright, 1951):

To allow comparisons with other work, the value of FST was also calculated by BIOSYS-1 (Swofford & Selander, 1981), using the WRIGHT78 step that corrects for sampling error.
The private allele method employs a relationship between the mean frequency of private alleles, ¯p (1), and the gene flow measured by Nm:

A correction was used for the mean sample size (NS) by multiplying Nm by 25/ NS (Slatkin, 1985; Barton & Slatkin, 1986). Calculations were computed using GENEPOP.
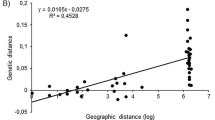
Isolation-by-distance was studied using a method adapted from Slatkin (1993), computing θ-values between all pairs of populations and studying the relationship between those values and the geographical distance between populations. The DIST program (Slatkin, 1993) was used to compute pairwise θ-values. The GENEPOP program was used to test independence between θ and the geographical distance using a Mantel's test.
Results
Among the 15 studied loci, seven were monomorphic at the 99% level (details of allele frequencies are available from the corresponding author). The seven polymorphic loci were Pgm, Mpi, Hbdh, Idh, Ak, Gpi and Acp. Sample sizes ranged from 219 to 24 for Pgm, 122 to 24 for Mpi, 166 to 24 for Hbdh, 220 to 24 for Idh, 140 to 24 for Gpi and 123 to 24 for Ak and Acp. Mean heterozygosity was 0.104 (SD=0.052). The loci Pgm, Mpi, Hbdh and Idh exhibit a private allele. All other alleles occur in both an African population and a European population.
Among 134 Fisher's tests carried out, only five (3.7%) showed a significant deviation of genotype frequencies from the Hardy–Weinberg expectations. Three of them concern Acp (among five populations in which this locus is polymorphic); the others concern Mpi for the Boisviel (BOI1) population and Ak for the Plateau de Lhers (PYR) population. Mean values of F and f across the seven loci are low: 0.046 (SD=0.029) and 0.053 (SD=0.026), respectively. On the other hand, values of F and f are high for Acp: 0.798 (SD=0.193) and 0.801 (SD=0.190), respectively. These values indicate a heterozygote deficiency. Examination of the genotypical frequencies revealed that three populations contained only homozygotes for Acp. Because of this important deviation, the Acp locus was excluded from further analysis. The loss of information was low considering the low polymorphism of this locus (the main allele accounts for 98.9% of scored alleles).
Table 3 presents the results of comparisons of allele frequencies across localities using Fisher's test. Significant differences exist for Pgm, Mpi, Hbdh and Gpi when comparing the 22 populations. The global test across loci is highly significant. The test across loci is not significant when comparing populations within the zones of northern Africa and western Africa, the only significant difference being for Mpi in northern Africa. On the other hand, significant differences exist for Pgm, Mpi, Gpi and across loci in the zone of France and Portugal. Results of comparisons of populations within France and Portugal are presented in Table 4. Comparison of populations within the south-east of France revealed no significant differences. Considering this lack of significant differentiation, these five populations were pooled for further comparisons to increase the power of the tests. There is a high level of heterogeneity: all pairs of populations exhibit significant differences in allele frequencies. Table 5 shows comparisons between homogeneous zones or regions after pooling of populations. There is no significant difference across loci between western Africa and northern Africa. On the other hand, allele frequencies for the south-east region of France are significantly different from both African zones.
θ-values calculated from Weir & Cockerham (1984) are presented in Table 6. The northern Africa zone was not studied here (only two populations were sampled). The values are low, whatever the geographical scale considered: less than 0.0098 at the regional scale, less than 0.0099 at the zonal scale. The negative value of θ noticed in the region of Bobo-Dioulasso (incompatible with the mathematical definition of FST) results from the calculation method (Swofford & Selander, 1981) and reveals an absence of differentiation between populations. The value of θ across the 22 populations is 0.007 (SD=0.0031). This value is close to the value of FST=0.008 computed by BIOSYS-1 for the same populations. The mean genetic distance is 0.002 (SD=0.004). All Nm-values are high and range from 25 to 156.
Values of Nm calculated using the private alleles method are lower than the values calculated from θ. All values are greater than 1, whatever the geographical scale: 19 for the 22 populations, 4 in western Africa and 73 in France and Portugal.
There is no relationship between the geographical distance between two populations and their common θ-value. The significance level of the Mantel's test is 0.163 for the 22 populations, 0.2884 for western Africa and 0.0887 for France and Portugal.
Discussion
Among 26 loci of an Australian population of H. armigera, Daly & Gregg (1985) found a mean heterozygosity of 0.113, similar to the value of 0.104 obtained here with 15 loci.
In Heliothis virescens (F.), Heckel (in Mallet et al., 1993) shows sex-linkage for the Acp locus. Such a sex-linkage in H. armigera could be responsible for the deviations from Hardy–Weinberg equilibrium noticed here. Because the sex of individuals was not scored, this hypothesis could not be verified.
Whatever the geographical area, genetic differentiation between populations within a region (less than 50 km in diameter) is low. In the south-east region of France, there was no significant difference in allele frequencies. In western Africa, allele frequencies were homogeneous even at the zonal scale. Low θ-values also indicate a high gene flow between populations within a region. The region, as defined here (50 km in diameter), seems to form a group of panmictic populations. This result differs from the conclusions of Korman et al. (1993). Studying the genetic variability of H. virescens (whose biology is very similar to H. armigera) from the southern U.S.A., these authors concluded that panmictic populations of this species spread over areas of less than 8 km in diameter.
Study of the differentiation of populations at the zonal scale revealed two situations. Within the France and Portugal zone, although Nm was high (25 or 73 depending on the method of calculation), many significant allele frequency differences exist between populations or regions. On the other hand, Nm is also high in western Africa (62 or 4, depending on the calculation method), but no significant allele frequency difference was noticed. Several other workers have pointed out significant allele frequency differences between populations subject to high gene flow. Korman et al. (1993) obtained significant differences in H. virescens with Nm of 135. Pashley et al. (1985), studying the genetic structure of populations in another migrant Noctuidae (Spodoptera frugiperda J. E. Smith), also found significant differences in allele frequencies for three loci out of eight, with a Nm of 7.6 (after removal of one population genetically very different from the others). For Buès et al. (1994), the existence of nonmigrants in a population of Agrotis ipsilon (Hufnagel) could explain the observed heterogeneity during some periods between subpopulations in Switzerland and in the south of France, despite high gene flow (Nm=35). Similarly for H. armigera in the France and Portugal zone, despite migrations through the Pyrenees (Anglade, 1969), the overwintering diapausing part of the population (Poitou & Buès, 1979) could be responsible for the observed genetic differentiation. On the other hand, in western Africa, the low genetic differentiation agrees with the hypothesis of a very small role for the diapause in the life history of H. armigera, whereas migration is of great importance. The annual cycle of extinction and recolonization experienced by western African populations could prevent differentiation by genetic drift and could be responsible for the observed lack of genetic structure (Slatkin, 1987).
The FST-value calculated from Daly & Gregg's (1985) data (12 Australian H. armigera populations sampled in a 3000-km-wide area) is 0.012 (Nm=21). This value has the same magnitude as that in the present study. The mean genetic distance (Nei, 1978) calculated by the same authors is 0.004 (SD=0.002), which is similar to our 0.002.
Although there are significant differences in allele frequencies for four loci across our 22 populations, the θ-value was low (0.007) and, thus, the Nm-value was high (36), revealing high gene flow. The lack of isolation-by-distance, expressed by the lack of correlation between θ and geographical distance, strengthens the hypothesis of high gene flow among the 22 populations. Moreover, no significant difference was noticed in allele frequencies between populations located on either side of the Sahara desert. The existence of high gene flow between populations from northern and western Africa suggests that the Sahara desert is not an insurmountable barrier for migrating H. armigera. This hypothesis agrees with that of Hmimina et al. (1993) in Morocco following an ecophysiological comparison of H. armigera samples. Migration on such scales implies flights across minimum distances of 2000 km from the northern frontier of the Sahara to the sub-Saharan zone. Such long migrations were also suggested by the trapping of H. armigera moths on Ascension Island, 2000 km from the African coast (Bowden & Johnson, in Widmer & Schofield, 1983). The low number of migrants sufficient to overcome the effects of genetic drift (Spieth, 1974; Slatkin, 1987) suggests that the few moths able to survive a trans-Saharan migration could be responsible for the lack of genetic structure noticed. On the other hand, it is also possible that this lack of structure results from the limits of the methods used. These limits depend on the sampling of populations, the number of loci studied and the underestimation of enzyme polymorphism resulting from the lack of separating power of electrophoresis (Fergusson, 1980; Pasteur et al., 1987). It is also possible that the condition of gene neutrality was not met and that the lack of differentiation was caused by a uniform selection pressure among populations. Finally, it is possible that the low genetic variability could be the result of a recent introduction of H. armigera in the northern part of its distribution or in western Africa. A similar hypothesis was proposed by Gasperi et al. (1991) for Ceratitis capitata Wied. in the Mediterranean area and in Africa. Better knowledge of dispersal in H. armigera will require further studies of the genetic variation between populations originating from the whole distribution area of the species, using molecular or ecophysiological methods.
References
Anglade, P. (1969). Premières observations de déplacements orientés de Noctuelles et de Sphingides dans une haute vallée Pyrénéenne par recapture d'insectes marqués. Bull Soc Entomol Fr, 74: 59–63.
Barton, N. H. and Slatkin, M. (1986). A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity, 56: 409–415.
Bowden, J. (1973). Migration of pests in the tropics. Medelingen van de Faculteit Landbouwwetenschappen Rijksuniversiteit Gent, 38: 785–796.
Buès, R., Freuler, J., Toubon, J. F., Gerber, S. and Poitout, S. (1994). Stabilité du polymorphisme enzymatique dans les populations d’un Lépidoptère migrant, Agrotis ipsilon. Entomologia exp appl, 73: 187–191.
Cayrol, R., Poitout, S. and Anglade, P. (1974). Etude comparée des caractères biologiques respectifs de quelques espèces de Noctuidae plurivoltines migrantes et sédentaires. Ann Zool Ecol Anim, 6: 1–10.
Daly, J. C. and Gregg, P. (1985). Genetic variation in Heliothis in Australia: species identification and gene flow in two pest species H. armigera (Hübner) and H. punctigera Wallengren (Lepidoptera: Noctuidae). Bull ent Res, 75: 169–184.
Duviard, D. (1981). Les Dysdercus du Cotonnier en Afrique Occidentale. Ecologie et Migrations. Office de la Recherche Scientifique et Technique Outre-Mer, Paris.
Fergusson, A. (1980). Biochemical Systematics and Evolution. Blackie, Glasgow.
Fisher, R. A. (1938). Statistical Methods for Research Workers. 7th edn. Oliver and Boyd, Edinburgh.
Gasperi, G., Guglielmino, C. R., Malacrida, A. R. and Milani, R. (1991). Genetic variability and gene flow in geographical populations of Ceratitis capitata (Wied.) (medfly). Heredity, 67: 347–356.
Giret, M. and Couilloud, R. (1986). Substitution of agar-agar by a carraghenate based gel to make nutrient mediums for the rearing of Lepidoptera larvae. Cot Fibr Trop, 41: 131–133.
Hardwick, D. F. (1965). The corn earworm complex. Mem Ent Soc Can, 40: 1–247.
Hillis, D. M. and Moritz, C. (1990). Molecular Systematics. Sinauer Associates, Sunderland, MA.
Hmimina, M., Poitout, S. and Buès, R. (1993). Variabilité des potentialités diapausantes intra et interpopulations chez Heliothis armigera Hb (Lep: Noctuidae). J Appl Ent, 116: 273–283.
Korman, A. K., Mallet, J., Goodenough, J. L., Graves, J. G., Hayes, J. L., Hendricks, D. E. et al. (1993). Population structure in Heliothis virescens (Lepidoptera: Noctuidae): an estimate of gene flow. Ann Entomol Soc Am, 86: 182–188.
Mallet, J., Korman, A., Heckel, D. G. and King, P. (1993). Biochemical genetics of Heliothis and Helicoverpa (Lepidoptera: Noctuidae) and evidence for a founder event in Helicoverpa zea. Ann Entomol Soc Am, 86: 189–197.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics, 89: 583–590.
Nibouche, S. (1994). Cycle évolutif de Helicoverpa armigera (Hübner, 1808) (Lepidoptera, Noctuidae) dans l'ouest du Burkina Faso). Doctoral thesis, Ecole Nationale Supérieure Agronomique de Montpellier.
Pashley, D. P., Seth, J. J. and Spark, A. N. (1985). Genetic population structure of migratory moths: the Fall Armyworm (Lep. Noctuidae). Ann Entomol Soc Am, 78: 756–762.
Pasteur, N., Pasteur, G., Bonhomme, F., Catalan, J. and Britton-Davidian, J. (1987). Manuel Technique de Génétique par Électrophorèse des Protéines. Lavoisier, Paris.
Poitout, S. and Buès, R. (1974). Elevage de chenilles de vingt-huit espèces de lépidoptères Noctuidae et de deux espèces d'Arctiidae sur milieu artificiel simple. Particularités de l'élevage selon les espèces. Ann Zool Ecol Anim, 6: 431–441.
Poitout, S. and Buès, R. (1979). La noctuelle de la tomate (Heliothis ou Helicoverpa armigera Hbn.). Son cycle évolutif dans le sud de la France. Défense des Végétaux, 195: 12–28.
Raymond, M. and Rousset, F. (1995). GENEPOP Version 2). Institut des Sciences de l’Evolution, Montpellier.
Slatkin, M. (1985). Rare alleles as indicators of gene flow. Evolution, 39: 53–65.
Slatkin, M. (1987). Gene flow and the geographic structure of natural populations. Science, 236: 787–792.
Slatkin, M. (1993). Isolation by distance in equilibrium and non-equilibrium populations. Evolution, 47: 264–279.
Spieth, P. T. (1974). Gene flow and genetic differentiation. Genetics, 78: 961–965.
Swofford, D. L. and Selander, R. B. (1981). BIOSYS-1: a Fortran program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered, 72: 281–283.
Weir, B. S. (1990). Intraspecific differentiation. In: Hillis, D. M. and Moritz, C. (eds) Molecular Systematics pp. 373–410. Sinauer Associates, Sunderland, MA.
Weir, B. S. and Cockerham, C. C. (1984). Estimating F-statistics for the analysis of population structure. Evolution, 38: 1358–1370.
Widmer, M. W. and Schofield, P. (1983). Heliothis Dispersal and Migration). Tropical Development and Research Institute, London.
Wright, S. (1931). Evolution in Mendelian populations. Genetics, 16: 97–159.
Wright, S. (1951). The genetical structure of populations. Ann Eugen, 15: 323–354.
Acknowledgements
This work was a collaboration between the Institut d'Études et de Recherches Agricoles (INERA, Burkina Faso), the Montfavet Zoology Station of the Institut National de la Recherche Agronomique (INRA, France) and the Centre de Coopération Internationale en Recherche Agronomique pour le Developpement (CIRAD, France). Funding for this research was provided by INERA, INRA and CIRAD. We thank M. E. Hochberg for critical reading of the manuscript. Thanks are also due to Mrs Carola Meierrose (Evora University, Portugal), L. El Jadd (INRA, Afourer, Morocco), B. Chermiti (ESH, Sousse, Tunisia), T. Alaux and J. M. Vassal (IDESSA, CIRAD, Ivory Coast) for providing the insects. We also thank Mrs L. Boudinhon for her participation in electrophoresis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nibouche, S., Buès, R., Toubon, JF. et al. Allozyme polymorphism in the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae): comparison of African and European populations. Heredity 80, 438–445 (1998). https://doi.org/10.1046/j.1365-2540.1998.00273.x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1046/j.1365-2540.1998.00273.x
Keywords
This article is cited by
-
Intra species diversity of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) in relation to geography and host plants affiliation in Uttarakhand Himalayan population, India
Phytoparasitica (2022)
-
Helicoverpa armigera Harm 1 Haplotype Predominates in the Heliothinae (Lepidoptera: Noctuidae) Complex Infesting Tomato Crops in Brazil
Neotropical Entomology (2021)
-
Multiple incursion pathways for Helicoverpa armigera in Brazil show its genetic diversity spreading in a connected world
Scientific Reports (2019)
-
Mitochondrial DNA and trade data support multiple origins of Helicoverpa armigera (Lepidoptera, Noctuidae) in Brazil
Scientific Reports (2017)
-
Genetic hitchhiking and resistance evolution to transgenic Bt toxins: insights from the African stalk borer Busseola fusca (Noctuidae)
Heredity (2017)