Abstract
Segregation analysis is a method of detecting major genes for quantitative traits without using marker information. It serves as an important tool in helping investigators to plan further studies such as quantitative trait loci mapping or more sophisticated genomic analyses. However, current methods of segregation analysis for a single trait typically have low statistical power. We propose a multivariate segregation analysis (MSA) that takes advantage of the correlation structure of multiple quantitative traits to detect major genes. This method not only increases the statistical power, but allows dissection of the genetic architecture underlying the trait complex. In MSA the observed phenotypes of multiple correlated traits are fitted to a multivariate Gaussian mixture model. Model parameters are estimated under the maximum likelihood framework via the expectation-maximization algorithm. The presence of major genes is tested using likelihood ratio test statistics. Pleiotropy is distinguished from close linkage by comparing three possible models using the Bayesian information criterion. Two simulation experiments were performed based on the F2 mating design. In the first, the statistical properties of MSA under varying heritabilities and sample sizes were investigated and the results compared with those obtained from single-trait analysis. In the second simulation the efficacy of MSA in separating pleiotropy from close linkage was demonstrated. Finally, the new method was applied to real data and detected a major gene responsible for both plant height and tiller number in rice.
Similar content being viewed by others
Introduction
It has generally been thought that quantitative traits are controlled by an infinite number of loci, each with an infinitely small effect – the ‘infinitesimal’ model (Lynch and Walsh, 1998). This view has been challenged by the observation that many quantitative traits are controlled by a few major genes plus numerous genes with small effects – the ‘oligogenic’ model. Segregation analysis that identifies major genes solely on phenotypic information is the basis of quantitative trait loci (QTL) mapping, since the current statistical methods and sample sizes used in gene mapping studies can detect only genes with major or moderate effects. In the post-genomics era, segregation analysis serves as an important intermediate tool to help investigators plan more sophisticated genomic studies. It also enables plant breeders to manipulate major genes (Wang et al., 2001; Zeng and Li, 2003; Yang et al., 2006).
A number of statistical methods have been developed to detect major genes for a variety of species (Lynch and Walsh, 1998). The most widely accepted is segregation analysis based on the mixture model(Tan and Chang, 1972; Elston and Stewart, 1973; Tan and D'Angelo, 1979; Elston, 1984; Loisel et al., 1994; Zhang et al., 2003). Various analytical strategies have been adopted in an attempt to make the method more reliable and robust. A mixed-inheritance model that incorporates the polygenic effect has been developed (Wang and Gai, 1997; Wang et al., 2001). Gai and Wang (1998) also proposed joint segregation analysis to fit various mixed-inheritance models using phenotypic information from multiple populations. This method has been applied successfully to major gene detection in many plant species (Wang and Gai, 1997; Wang et al., 2001). Other approaches, such as complex segregation analysis (which takes advantage of transmission information from traits within pedigrees), have been widely used in humans and animals and have been extended to plant studies (Tourjee et al., 1995; Aulchenko et al., 1999). A major limitation of phenotype-based segregation analysis, however, is that it lacks sufficient statistical power to detect genes with relatively low heritability. Its power is also low in more complex inheritance models owing to the increased number of unknown parameters.
In real experiments it is common to score a number of traits simultaneously. These traits are often correlated through a common genetic basis, such as pleiotropy or close linkage, or through shared environmental influences. Joint analysis of multiple traits can yield more substantial benefits than can be achieved through single-trait analysis. Statistically, multivariate analysis allows information from correlations between variables to be incorporated, which should result in increased statistical power and more precise estimates. Moreover, the increase in dimension improves the degree of separation between mixture components, further augmenting its power (Hamilton, 1991; Dolan and van der Maas, 1998). Biologically, multivariate analysis is likely to yield more interesting and informative results with respect to the interplay among the trait complexes. Multitrait genetic analysis has been used routinely in human genetics to improve the efficacy of gene detection in complex diseases (Blangero and Konigsberg, 1991; Almasy et al., 1997; Schmitz et al., 1998; Chien et al., 2006) but has not yet been applied to analysis in line crosses.
The objective of this study was to develop a means of segregation analysis that would use multiple correlated traits in line-crossing experiments – ‘multivariate segregation analysis’ (MSA). Observed phenotypes of multiple correlated traits were fitted to a multivariate Gaussian mixture model. Parameter estimations were obtained using the maximum likelihood (ML) method, implemented via the expectation-maximization (EM) algorithm (Dempster et al., 1977). The presence of major genes was tested using the likelihood ratio test (LRT) statistic. A test to determine whether the significant effects were due to pleiotropy or close linkage was also performed. Simulations under a variety of scenarios were conducted to compare the results obtained using MSA with those produced by conventional single-trait analyses. The use of MSA with real data was demonstrated by testing rice for two traits: plant height and tiller number.
Theory and methods
Genetic model
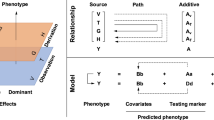
Let A and a be the two alleles of the locus controlling k correlated quantitative traits. In the segregating population, the major gene will have three genotypes: AA, Aa and aa. Let  be a row vector of phenotypic values for the k traits measured from the jth individual in the segregating population. The phenotypic values may be described by the following linear model
be a row vector of phenotypic values for the k traits measured from the jth individual in the segregating population. The phenotypic values may be described by the following linear model

where Xj=(1 X1j X2j) is the indicator variable vector for the genotype Gj of the jth individual with Xj=H1=(1 1 0) when Gj=AA, Xj=H2=(1 0 1) when Gj=Aa, and Xj=H3=(1 −1 0) when Gj=aa. Let

be the matrix of parameters, where b01, b02, ċ, b0k correspond to the population mean of the k traits, b11, b12, ċ, b1k and b21, b22, ċ, b2k correspond to the additive effects and dominance effects, respectively, for the major gene. Let  be the vector of residual effects with an assumed multivariate Gaussian distribution, that is, MVN(0, Ve), where
be the vector of residual effects with an assumed multivariate Gaussian distribution, that is, MVN(0, Ve), where  is a row vector of zeros and
is a row vector of zeros and

is the residual variance-covariance matrix.
Model (1) is now considered as a general multivariate linear model with values of Xj missing. Conditioning on the major gene genotype Gj, Yj has a density of  or
or  .
.
Multivariate segregation analysis
Since genotypes of the major gene in the segregating population are unknown, the total phenotypic value Yj of the jth individual for the k traits is a mixture of three Gaussian distributions, with mixing proportions equal to the genotype probabilities, denoted by p1j, p2j and p3j for the three possible genotypes AA, Aa and aa. The probability density of Yj is

where

is the probability density function of the jth individual conditioning on the lth genotype. The overall likelihood of n independent observations in the segregating population is

The ML estimates of parameters b and Ve in model (3) can be achieved using the EM algorithm (Dempster et al., 1977). It is easier to deal with the log-likelihood function, as shown below

The ML estimates of the unknown parameters can be found via the EM algorithm

and

The estimates in the above two formulae are the corresponding expectations of the missing variables. The final step is to obtain these expectations.
The prior probabilities of genotypes for the major gene depend on the type of population. For instance,  for an F2 population and
for an F2 population and  for a backcross population BC1=F1 × P1. According to Bayes' theorem, the posterior probability for each of the major genotypes plj* is
for a backcross population BC1=F1 × P1. According to Bayes' theorem, the posterior probability for each of the major genotypes plj* is

The posterior probabilities can be used to calculate the expectations in Equations (5) and (6), as shown below

The iteration process of the EM algorithm is summarized as follows
-
1)
Choose initial values of the parameters b(0) and Ve(0).
-
2)
E-step: Calculate the posterior probabilities for all the possible genotypes for the jth individual using Equation (7) and the expectations using Equation (8).
-
3)
M-step: Calculate the estimates of parameters b(1) and Ve(1) using Equations (5) and (6). The results are then used to update the initial values.
-
4)
Repeat the E-step and the M-step until the iterations converge.
The proportions of the three major gene genotypes in a segregating population at the tth iteration are calculated using

Tests of major genes
For the sake of demonstration, here we restrict our discussion to bivariate segregation analysis in an F2 population. Five hypotheses regarding the general and restricted models are considered.
H1: No major gene is segregating in the population, that is, b11=b12=b21=b22=0. The parameters to be estimated are b=(b01 b02) and

The ML estimates of the parameters under this hypothesis are

Define the log-likelihood function evaluated at the solutions as

where  .
.
H2: There is a major gene segregating in the population. The model under this hypothesis is the same as model (1). The ML estimates of parameters b and Ve are obtained by applying Equations (5) and (6). The log-likelihood function under this hypothesis is given in Equation (4) and denoted by ln L2 here.
H3: There is a mixture of three multivariate normal distributions with a common covariance matrix. The model under this hypothesis is similar to the single-gene model except that the proportions of the three normal distributions pl (l=1, 2, 3) are not fixed. The ML estimates of the corresponding parameters at convergence are denoted by  and
and  , respectively. Denote the log-likelihood value at convergence by
, respectively. Denote the log-likelihood value at convergence by  , where
, where  .
.
H4: The major gene regulates the first trait only, that is, b21=b22=0, the parameters to be estimated are b01, b02, b11, b12 and Ve. Under this model, the log-likelihood function evaluated at the solutions is denoted by ln L4.
H5: The major gene regulates the second trait only. Here the model is restricted to b11=b12=0, with the parameters to be estimated being b01, b02, b21, b22 and Ve. The log-likelihood function evaluated at the model solutions is denoted by ln L5.
The likelihood-ratio test (LRT) is employed to compare the hierarchical models based on the hypotheses stated above, that is, to evaluate whether a reduced model gives essentially the same fit as the general model.
The LRT statistic for testing the existence of a major gene is given by

The test statistic asymptotically follows a chi-square distribution with 2k degrees of freedom.
The LRT statistic that tests whether the pattern of distribution is consistent with a single-gene model is calculated based on hypothesis H2 and its reference hypothesis H3. Therefore, hypothesis H2 is nested within the general model under H3 with the additional constraints  . The test statistic will be
. The test statistic will be 
Once a major gene is detected using LR2, we can determine whether it has pleiotropic effects on both traits. The test is then equivalent to the tests for trait-specific effects. Rejection of both trait-specific hypotheses indicates the presence of pleiotropic effects. The LRT statistics are therefore calculated as  and
and  .
.
The test statistics LR2, LR3 and LR4 each will follow approximately a chi-square distribution with two degrees of freedom under the corresponding null hypotheses, since there are two additional free parameters being fit under the general model. Once a single-gene model is rejected, we need to consider more complex models containing several major genes, which will not be discussed in this paper.
Pleiotropy versus close linkage
Both pleiotropy and linkage can contribute to the genetic correlation of different traits. Rejecting the last two hypotheses in the previous section does not distinguish whether the significant effect is due to one shared major gene or to multiple, closely linked genes, each with a major effect on a single trait. Further separation of these two possibilities may be intrinsically important in practical breeding or other genetic studies. To this end, MSA can be extended to consider the linkage model. In this study we confine our consideration to the hypothesis of two traits regulated respectively by two genes without pleiotropies. Let r denote the recombination fraction between the two linked genes. Clearly, r=0 infers that there is only one major gene, which points to a pleiotropic model, and r=0.5 can be interpreted as two unlinked genes, indicating a non-linkage model.
The density of phenotypic values under the close linkage model (r takes value between 0 and 0.5) is analogous to that in Equation (2), except that the mixture now has nine components, since recombination results in nine possible joint genotypes for two genes in an F2 mating design. Accordingly, the likelihood function should be reconstructed. The ML estimates of the parameters, including the additional unknown r, can be obtained by following the EM procedure described above, but with some modifications:
In the initial step, a starting value also has to be manually assigned to r, denoted by r(0).
In E-step, the posterior probabilities of nine genotypes are calculated according to Bayes' theorem, where the priors depend on the recombination fraction between two genes;
In M-step, r can be estimated by the ML method based on a multinomial distribution model, the estimate of which is then taken as the initial value of the next sweep.
The test of pleiotropy versus linkage is implemented by choosing the most suitable model among the three candidates, namely, the linkage model in which r is estimated, the complete pleiotropic model (r is constrained to 0, i.e., one major gene model) and the non-linkage model (r is constrained to 0.5). Here we propose to use the Bayesian information criterion (BIC) that does not assume nesting of models as the selection criteria (Schwarz, 1978). The model with the smallest BIC will be chosen. The BIC is defined as follows

Data analysis
Simulation A
The main purpose of this simulation was to explore the statistical properties of MSA under varying conditions and compare them with those of conventional single-trait analysis. To do so, we simulated three correlated traits under the F2 mating design. Three levels of h2 were simulated for the major gene: 30, 50 and 70%, each under two different sample sizes (n): 200 and 300. (See Table 2 for a complete list of the scenarios.) Two hundred replicated simulations were conducted for each of the six scenarios. Under each scenario, two simulation designs were considered. In the first, the three traits were assumed to be affected by a single gene. The additive effects of the gene on the three traits were 0.5, 1.0 and −0.5, respectively, whereas the dominance effects were set to be 1.0, 0.5 and 1.0, respectively. The residual correlation coefficients were −0.5, 0 and 0.5 between traits 1 and 2, 1 and 3, and 2 and 3. The residual variances of the traits were chosen based on the heritabilities and genetic variances of the major gene. The residual covariance was determined by the corresponding residual correlation coefficient and the residual variance. The population means for the three traits were set to be b01=10, b02=20 and b03=15. In the second design, a major gene was assumed to control the second trait only (no pleiotropic effect on the other traits). In this case, the phenotypic correlation was solely due to the shared environmental influences. Such a model is represented by b11=b21=b13=b23=0 where the phenotypic variances of the first and third traits are completely due to the residual variances. The values of the residual variances and covariances are given in Table 1. For comparison, each data set was analyzed with univariate, bivariate and trivariate segregation methods. The following criteria were used to evaluate the methods:
-
1)
statistical power for major gene detection, determined by the proportion of the 200 replicates in which the major gene was detected;
-
2)
precision and accuracy of the estimated effects of the major gene and the estimated residual variances and covariances.
The empirical statistical powers from seven segregation methods are summarized in Table 2, from which we can draw the following conclusions. First, as would be expected, statistical power is enhanced with increased assample size and heritability, although it is more sensitive to changes in the latter. Second, MSA has a substantially higher power than univariate analysis in design I. Even in the case with the strictest conditions, the power of the trivariate segregation method reached 100%, compared with around 10% in conventional single-trait analyses. Powers are also significantly improved in the bivariate cases. The joint analysis of traits 1 and 3, however, tended to produce relatively lower power, an effect that may be caused by the non-correlated residual variances of the two traits. Third, MSA can improve power even in univariate cases where traits have no shared major gene. It can be seen that in design II, under the first two treatments powers can be improved by up to around twofold with bivariate analyses, and up to fourfold with trivariate analyses. This result further demonstrates the effectiveness of both genetic and shared environmental correlations between traits as aids to the detection of major genes.
Figure 1 shows the means and standard deviations (s.d.) of the estimated parameters from the MSAs and the univariate analysis under the two simulation designs. The results showed the expected trend: higher heritability and larger sample size tend to produce more accurate and precise estimates. In the first simulation design, the multivariate methods significantly improved the performance of segregation analysis by showing that all estimates are close to the true values. We can also see that the trivariate method produced much more precise results than the other two methods (indicated by the short s.d. bars). However, the bivariate method showed no apparent advantage in terms of precision. In the second design (with no shared gene), all three multivariate methods yielded better results than the single-trait method, with the exception of the bivariate analysis of traits 1 and 3 (M-13), which would be expected since there is neither a genetic nor an environmental correlation between the two traits in that case.
Comparison of the means and standard deviations (s.d.) of estimated genetic parameters from the trivariate (M-123), bivariate (M-12; M-13; M-23) and univariate methods (S-1; S-2; S-3) under different scenarios of the simulation experiment. (a) and (b) are for the first and second designs of simulation, respectively. The vertical bars indicate±s.d.
Simulation B
In this simulation, we focused on the testing of close linkage and pleiotropy. Two major genes were assumed to control two traits independently, with the recombination fraction between the genes (r) varying under six levels from 0 to 0.5. The heritability for each major gene was simulated under two levels: 50 and 70%. The sample size was set at 300. The genetic effects of genes, residual correlations between traits and other parameters were set to be the same as those of the first two traits in simulation A.
We have therefore created 12 different simulation scenarios. Each dataset generated was then analyzed using the procedure described above. The resulting frequencies of acceptance of the candidate models over 100 replicates are presented in Table 3, where the complete pleiotropic model is referred to as Model I, the linkage model as Model II and the non-linkage/independent model as Model III. We see from the table that the method has extremely high power to accept the true model under two extreme scenarios, when r equals 0 and 0.5, which means that instances of one shared major gene and two unlinked genes can be detected without difficulty. But when r=0.4, the frequency of identification of Model II is much lower, even under the high heritability of 70%. This is understandable since the simulated two genes are very loosely linked, making it difficult to distinguish between Models II and III with only phenotypic information. This particular result is not important, however, since we are usually more interested in separating close linkage from pleiotropy, rather than loose linkage from non-linkage. It is encouraging to find that our method has satisfactory powers to detect Model II under scenarios where r equals 0.1, 0.2 and 0.3. The powers are equal or very close to 100% when h2=70%. In cases with low heritability, a clear trend emerged in which shorter distances between genes resulted in greater frequencies of acceptance of the false model of pleiotropy. Simulations indicate that a paradoxical result could be obtained when the recombination fraction is smaller than 0.1. To test genes with shorter distances, sample size can be increased or information incorporated from sources beyond phenotypes. Means and standard deviations of the estimates of r and the genetic effects obtained from the true model are also given in Table 3. All estimates seem to be close to the true values and have good precisions except the cases when r=0.4, where the estimates of recombination fraction are relatively biased. In general, the scenarios with high heritabilities produced greater accuracy and precision.
Example of real data analysis
We used real data collected from a rice experiment to illustrate the proposed MSA method for major gene detection. An F2 population of 597 plants was derived from a cross between Duonieai and Zhonghua 11. Two agronomic traits were examined: plant height and tiller number. The joint frequency distributions of the two traits are given in Table 4, clearly showing a degree of correlation. The data were fitted into the multivariate model. The results of hypothesis testing are listed in Table 5, from which we can see that:
-
1)
the LR value for testing the presence of a major gene is significant at 1% level, rejecting H1;
-
2)
the LR value for testing the presence of a single major gene influencing the variation of the traits is not significant, and thus we accept H2;
-
3)
both H4 and H5 are rejected by the LR tests.
The test results provide evidence showing the existence of a single major gene with pleiotropic effects on both traits. The additive and dominance effects of the major gene on plant height are −21.3 and 40.6 cm, respectively, indicating overdominance. For tiller number, the additive and dominance effects are 22.7 and −25.3, respectively, near complete dominance. Estimates of the genetic correlation coefficient and residual correlation coefficient between the two traits are −0.97 and −0.28, respectively. The high genetic correlation coefficient between the two traits further demonstrates the pleiotropic mechanism of the major gene. Figure 2 depicts the joint distribution of the two traits when the parameters are estimated under the H2 hypothesis, that is, when the constraint (p1j=0.25, p2j=0.5, p3j=0.25) is enforced. Although the effect was significant these results did not exclude the possibility of close linkage, where two independent genes determine different traits. We therefore performed the proposed procedures to test pleiotropy versus linkage. The results from fitting the three possible models are presented in Table 6. The pleiotropic model was chosen as the most suitable model because it had the smallest BIC, further supporting the hypothesis that plant height and tiller number in rice are under the control of one major gene. Our analysis therefore suggests that future molecular mapping of genes influencing these two traits or other functional analyses will have a high probability of success if conducted using the same population.
Discussion
Segregation analysis of phenotypic data is designed for preliminary screening of genes, and, while it provides estimates of heritability and related parameters, it furnishes no information on the exact locations of genes. It does supply evidence of genetic effects that would warrant gene mapping, whereas raditional statistical programs and sparse genetic markers are not sufficient to tackle a trait with no major gene effects. However, the conventional method of segregation analysis usually has insufficient power, especially in cases with low heritability and complex inheritance modes. In this study, we have shown that MSA has advantages not offered by single-trait segregation analysis. Results of the simulations demonstrate that if a major gene has a pleiotropic effect on both traits, MSA can have significantly higher power than univariate segregation analysis, with substantial improvements in the precision and accuracy of parameter estimation. The gain in power was also observed when the major gene has sole regulation of one trait, suggesting that shared environmental correlation between traits can effectively aid the detection of genes with large effects. We have demonstrated that even without marker information, MSA can effectively test for pleiotropy versus close linkage. The results will facilitate our understanding of the genetic architecture underlying trait complexes, as well as providing valuable guidance for practical breeding, where breaking unfavorable gene linkages is an important goal.
Substantial work has been done on joint mapping for multiple quantitative traits. Jiang and Zeng (1995) carried out seminal work in extending composite interval mapping to a multivariate method. Xu et al. (2005) made the procedure applicable to multiple discrete traits. Methods have also been developed to handle longitudinal data by fitting them to a particular growth trajectory (Wu et al., 2004; Yang et al., 2006). The proposed method presented in this paper is similar to multivariate QTL mapping, although there are further limitations that need to be considered in segregation analysis. In MSA, the genotypes of putative loci cannot be inferred from flanking markers and, clearly, no marker effects can be fitted into the model for genetic background control. A noteworthy difference between the method proposed by Jiang and Zeng (1995) and ours is the means of testing pleiotropy and close linkage. Jiang and Zeng used an optimized two-step strategy in which separate mappings of different traits were initially performed. This was followed by a two-dimensional search for maximum likelihood under the linkage model. The tests of hypotheses were then concentrated in the region near the peak indicated by the separate mappings. This strategy cannot be used in segregation analysis because of the difficulty of controlling the positions of genes. We tackled this problem by introducing an unknown variable r to measure the distance between genes. Pleiotropy and linkage were tested by comparing three models in which r is set at 0, 0.5 and unknown, respectively. The success of this strategy has been demonstrated by our simulations.
Dissection of the genetic architecture underlying the correlations among traits is an important goal of evolutionary biology. Genetic correlations can strongly affect the evolutionary trajectory of complex trait structures because natural selection of one trait could cause an evolutionary change in a correlated neutral character or alter the response to selection in a correlated trait that is under direct selection (Conner, 2002). In other words, pleiotropic connections between traits might cause compromises among adaptations of different traits and make them evolve in correlated patterns. However, correlations do not always put constraints on the joint evolution of the traits. A positive genetic correlation could facilitate adaptive evolution, given that positive directional selection occurs in all traits (Boake, 1994). Certain combinations of traits rarely occur randomly: they may be selected for their coordinated function or have a common genetic basis such as pleiotropy (Mclellan, 2005). Trait integration produced by a shared genetic basis is usually independent of the environment, whereas integration caused by functional association may vary across environments (Young and Badyaev, 2006). However, it cannot be concluded that traits evolve independently if only residual correlation is detected in a single experiment. There is a need for more experiments conducted in similar macro-environments (i.e., with the same regional or climatic conditions) to eliminate the possibility of functional integration. Understanding the genetic basis of correlations not only allows us to draw inferences about the influence of past selection on traits, but also enables us to make predictions about the hypothetical patterns of future selection. Moreover, knowledge of integration among traits is valuable in the recognition of species and the determination of relationships among taxa.
In this study, a method of detecting major genes and testing for pleiotropy versus linkage was demonstrated, based on cases with three traits and two traits, respectively. In real data analysis, more traits are recorded, and several factors may constrain the number of traits included in MSA. As this number increases so will the number of the generated hypotheses, which might become unmanageable in terms of interpretation and computational time. In addition, there is a need for more intermediate models in the test of pleiotropy versus linkage, for example, where one or more genes has pleiotropic effects on some traits but not others. Finally, the number of traits should be limited by the sample size to avoid increasing the parameters and thereby losing power. Similar issues have been raised and discussed in other studies (Schmitz et al., 1998; Knott and Haley, 2000). Investigators need to design multivariate analyses with care, choosing a reasonable subset of traits based on theoretical sense and intuitive knowledge of the relationships among traits. An alternative way of approaching multiple traits is to use dimension reduction strategies such as the principal component analysis (Korol et al., 1995; Mangin et al., 1998), where the data are transformed into fewer variables, called ‘super traits’, which account for the majority of the variation in the entire set of traits. After transformation, existing single-trait methods can be used directly. However, the results of super-trait analyses are often difficult to interpret in a biological setting (Hackett etal., 2001; Xu et al., 2005).
In summary, the present study has successfully demonstrated a powerful multivariate approach for major gene detection. As it was designed principally to test the applicability of the new multivariate strategy, the model was based on two traits controlled by a single major gene. This work can readily be extended to more major genes and to polygenic effects to increase its biological relevance. Further research is needed to develop more sophisticated models that use phenotypic information from multiple generations. Finally, MSA is not restricted to multiple traits. It can be used to perform repeated measurements of the same trait or analyses of longitudinal traits. The high statistical power of the MSA approach opens up a new opportunity to unravel the genetic architecture of multiple quantitative traits.
References
Almasy L, Dyer TD, Blangero J (1997). Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol 14: 953–958.
Aulchenko YS, Veprev SG, Axenovich TI (1999). An example of complex segregation analysis of plant pedigree: reversion of cytoplasm type in Sugar Beet (Beta vulgaris L.). Ann Hum Genet 63: 351–353.
Blangero J, Konigsberg LW (1991). Multivariate segregation analysis using the mixed model. Genet Epidemiol 8: 299–316.
Boake CRB (ed). (1994). Quantitative Genetic Studies of Behavioral Evolution. University of Chicago Press: Chicago, IL.
Chien KL, Chen WJ, Hsu HC, Su TC, Chen MF, Lee YT (2006). Segregation analysis of apolipoprotein A1 levels in families of adolescents: a community-based study in Taiwan. BMC Genet 7: 4.
Conner JK (2002). Genetic mechanisms of floral trait correlations in a natural population. Nature 420: 407–410.
Dempster AP, Laird NM, Rubin DB (1977). Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Ser B 39: 1–38.
Dolan CV, van der Maas HJL (1998). Fitting multivariate normal mixtures subject to structural equation modeling. Psychometrika 63: 227–253.
Elston RC (1984). The genetic analysis of quantitative trait differences between two homozygous lines. Genetics 108: 733–744.
Elston RC, Stewart J (1973). The analysis of quantitative traits for simple genetic models from parental, F1 and backcross data. Genetics 73: 695–711.
Gai JY, Wang JK (1998). Identification of major gene and polygene mixed inheritance model from backcrosses or F2:3 families. Acta Agron Sinica 24: 402–409.
Hackett CA, Meyer RC, Thoma WTB (2001). Multi-trait QTL mapping in barley using multivariate regression. Genet Res 77: 95–106.
Hamilton JD (1991). A quasi-Bayesian approach to estimating parameters for mixtures of normal distributions. J Bus Econ Statist 9: 27–39.
Jiang C, Zeng ZB (1995). Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140: 1111–1127.
Knott SA, Haley CS (2000). Multitrait least squares for quantitative trait loci detection. Genetics 156: 899–911.
Korol AB, Ronin YI, Kirzhner VM (1995). Interval mapping of quantitative trait loci employing correlated trait complexes. Genetics 140: 1137–1147.
Loisel R, Goffinet B, Monod H, de Oca GM (1994). Detecting a major gene in an F2 population. Bioimetrics 50: 512–516.
Lynch M, Walsh B (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates: Sunderland, MA.
Mangin B, Thoquet P, Grimsley N (1998). Pleiotropic QTL analysis. Biometrics 54: 88–99.
Mclellan T (2005). Correlated evolution of leaf shape and trichomes in Begonia dregei (Begoniaceae). Am J Botany 92: 1616–1623.
Schmitz S, Cherny SS, Fulker DW (1998). Increase in power through multivariate analyses. Behav Genet 28: 357–363.
Schwarz G (1978). Estimating the dimension of a model. Ann Statist 6: 461–464.
Tan WY, Chang WC (1972). Convolution approach to genetic analysis of quantitative characters of self-fertilized populations. Biometrics 28: 1073–1090.
Tan WY, D'Angelo H (1979). Statistical analysis of joint effects of major genes and polygenes in quantitative genetics. Biom J 21: 179–192.
Tourjee KR, Harding J, Byrne TG (1995). Complex segregation analysis of Gerbera flower color. Heredity 74: 303–310.
Wang J, Podlich DW, Cooper M, DeLacy IH (2001). Power of the joint segregation analysis method for testing mixed major gene and polygene inheritance models of quantitative traits. Theor Appl Genet 103: 804–816.
Wang JK, Gai JY (1997). Identification of major gene and polygene mixed inheritance model and estimation of genetic parameters of a quantitative trait from F2 progeny. Acta Genet Sinica 24: 432–440.
Wu RL, Ma CX, Lin M, Casella G (2004). A general framework for analyzing the genetic architecture of developmental characteristics. Genetics 166: 1541–1551.
Xu C, Li Z, Xu S (2005). Joint mapping of quantitative trait loci for multiple binary characters. Genetics 169: 1045–1059.
Yang R, Tian Q, Xu S (2006). Mapping quantitative trait loci for longitudinal traits in line crosses. Genetics 173: 2339–2356.
Young RL, Badyaev AV (2006). Evolutionary persistence of phenotypic integration: influence of developmental and functional relationships on complex trait evolution. Evolution 60: 1291–1299.
Zeng W, Li BL (2003). Simple tests for detecting segregation of major genes with phenotypic data from a diallel mating. For Sci 49: 268–278.
Zhang YM, Gai JY, Yang YH (2003). The EIM algorithm in the joint segregation analysis of quantitative traits. Genet Res 81: 157–163.
Acknowledgements
We are grateful to the editor and four anonymous reviewers for constructive comments that significantly improved this manuscript. This work was supported by the National Basic Research Program of China (Grant No. 2006CB101700), the National Natural Science Foundation of China (Grant No. 30370758) and the Program for New Century Excellent Talents in University, Ministry of Education of China, to CX.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xiao, J., Wang, X., Hu, Z. et al. Multivariate segregation analysis for quantitative traits in line crosses. Heredity 98, 427–435 (2007). https://doi.org/10.1038/sj.hdy.6800960
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.hdy.6800960
Keywords
This article is cited by
-
Growth, seed development and genetic analysis in wild type and Def mutant of Pisum sativum L
BMC Research Notes (2011)
-
Latent common genetic components of obesity traits
International Journal of Obesity (2008)