Abstract
Recently, a LAP protein, scribble, was identified in Drosophila epithelia as a basolateral protein that controls the apical-basolateral polarity. Loss of scribble causes disorganisation and overgrowth of the epithelia. Scribble has a human homologue, human scribble (hScrib), which is a substrate of ubiquitin-mediated degradation by human papillomavirus E6 and the E6AP ubiquitin-protein ligase. In the present study, we revealed that hScrib localised to the basolateral regions of the epithelial cell line MDCK and human uterine cervical epithelial tissues by immunofluorescence. Human scribble colocalised rather with the adherens junction protein E-cadherin, but not with the tight junction protein ZO-1. Histochemical analysis showed a dramatic decrease in the expression of hScrib with the progression of disease from normal uterine cervical tissues to invasive cervical cancers through the precursor lesions. In contrast, the expression of hScrib was retained in the throughout epithelial layer of the HPV-negative cervical high-grade squamous intraepithelial lesions (H-SIL). Although quantitative RT–PCR revealed no significant downregulation of hScrib mRNA expression in the H-SIL, it revealed a clear downregulation in the invasive cancers. These results suggest the possibility that degradation by HPV E6 is one of the causal roles for the progressive decrease of hScrib expression during the disease progression from low-grade squamous intraepithelial lesions to H-SIL, and a cooperative role of downregulation of hScrib mRNA expression and ubiquitin-mediated degradation of hScrib by E6 and E6AP led to the complete decrease of hScrib expression during the process of carcinogenesis from H-SIL to invasive cancer. These data underscore the importance of hScrib in the construction of tissue architecture and prevention of cancer development.
Similar content being viewed by others
Main
The organised polarity of cells is a hallmark of the epithelia. In cancer cells, the apical-basolateral polarity is disrupted and mislocalisation of many junctional proteins or hormone receptors is reported. Recently, Bilder et al (Bilder and Perrimon, 2000) identified the basolateral protein scribble that determines the localisation of the apical epithelial determinants. Loss of scribble mutation leads to loss of apical-basolateral polarity and the apical determinant Crumbs shows unrestricted localisation not just at the apical region in scribble mutants. scribble is localised at the septate junction, which is the analogue of the vertebrate tight junction. In scribble mutants, the adherens junctions are misdistributed around the cell periphery (Bilder and Perrimon, 2000). How do the basolateral proteins determine the localisation of the apical determinants at the apical region of cells and the precise localisation of the adherens junctions just above the septate junctions? Looking at its amino-acid sequence, scribble has four PDZ domains (named for its occurrence in the PSD-95, Discs-Large, and ZO-1 proteins), and it could have four different binding partners through four PDZ domains simultaneously. In addition, scribble has 16 canonical leucine-rich repeats (LRRs), another protein–protein interaction motif. Related repeats to the LRRs are known to bind small GTPases of the Ras superfamily (Jou and Nelson, 1998; Kaibuchi et al, 1999). Recently, proteins with both PDZ domains and LRRs have been reported to have common functions in the control of the cell shape and polarity. These proteins are grouped as LAP (LRR and PDZ domain) proteins (Bilder et al, 2000a). Scribble is supposed to function as a scaffold protein using these protein-binding motifs. Loss of scribble mutation also leads to the overgrowth of imaginal discs, follicle epithelia, and larval brain (Bilder et al, 2000b). Controlling the cell apical-basolateral polarity is closely related to the growth control (Greaves, 2000). So, scribble is not only an apical-basolateral polarity determinant but also a tumour suppressor.
Drosophila scribble has a human homologue, human scribble (hScrib), which was identified as a substrate of ubiquitin-mediated degradation by high-risk human papillomavirus (HPV) E6 and the E6AP ubiquitin-protein ligase (Nakagawa and Huibregtse, 2000). Tumour suppressor p53 is targeted for ubiquitin-mediated degradation by E6 dependent on the E6AP (Huibregtse et al, 1993; Scheffner et al, 1993, 1994; Huibregtse et al, 1995). P53 had a key role in the control of cell cycle and apoptosis (Kuerbitz et al, 1992; Lowe et al, 1993). By degrading p53, E6 disrupts the self-defending machinery against DNA damages of the host cells. Recently, it has been shown that the E6 targets not only the nuclear substrate but also the cytoplasmic substrates such as hScrib via ubiquitin-mediated pathway (Kiyono et al, 1997; Nakagawa and Huibregtse, 2000; Mantovani et al, 2001). The degradation of hScrib by E6 and E6AP was shown in the in vitro setting (Nakagawa and Huibregtse, 2000).
We sought to address two issues in this study. The first issue is whether hScrib shows basolateral localisation analogous to that of Drosophila scribble. The Drosophila scribble localises at the septate junction, which is functionally analogous to the vertebrate tight junction. We analysed localisation of hScrib on the subject of the localisation of other junctional proteins in the MDCK cell line and primary human cervical epithelia. Carcinogenesis of the uterine cervix has multistages from normal cervical epithelia to the invasive carcinoma through the squamous intraepithelial lesions. The second issue that we sought to address is whether hScrib is involved in the carcinogenesis of the uterine cervix. If so, the localisation of hScrib could be disrupted in cancer tissues and its expression is supposed to decrease with the progression of diseases, possibly depending on the E6 and E6AP. These issues would be discussed in the present study.
Materials and methods
Construction of the anti-hScrib antibodies
The DNA sequence, which encodes C-terminus (amino acids 1208–1632), was subcloned into the pGEX-6P-1 vector (Amersham Pharmacia, Buckinghamshire, UK). Glutathione S-transferase (GST) fusion protein, GST-hScrib C-terminus, was made in the bacteria and purified according to the manufacturer's recommendation. The amino acids encoding hScrib C-terminus were cleaved from the GST-fusion proteins with the PreScission Protease (Amersham Pharmacia, Buckinghamshire, UK) and purified. This hScrib C-terminus was injected into rabbits as antigens. The hScrib C-terminus antibody (anti-hScrib C-terminus) was purified from the serum of the immunised rabbits by affinity chromatography.
Western blotting
293 T cells were grown in DMEM medium supplemented with 10% foetal bovine serum. Protein extracts of 293 T cells, primary cervical tissues, and normal control tissues were made in the NP-40 lysis buffer containing 100 mM Tris (pH 8.0), 100 mM NaCl, and 1% NP-40. For primary tissues, cryostat sections were cut for haematoxylin and eosin staining to confirm the spread of the neoplastic lesions, and non-neoplastic tissues were trimmed off from the frozen tissues. For the normal cervix samples, we also trimmed off the underlying connective tissues. Protein concentration was determined by standard Bradford assay. Equal amounts of extracts were fractionated by SDS–PAGE and electrophoretically transferred onto the polyvinylidene difluoride membranes (Millipore Co., Bedford, MA, USA). The anti-hScrib C-terminus antibody (anti-hScrib C-terminus) was used at the dilution of 1 : 1000 to detect the expression of hScrib as indicated. The in vitro-translated hScrib and human homologue of Drosophila Discs-Large using the reticulocyte lysate system (Promega Co., Madison, WI, USA) were used as the positive and negative control, respectively. The level of protein expression was analysed by the STORM 860 according to the manufacturer's recommendation (Molecular Dynamics).
Immunofluorescence of MDCK cells
Subconfluent MDCK cells were grown on coverslips in the culture medium. Cells were washed three times with phosphate-buffered saline (PBS) and then fixed with 3.7% formaldehyde in PBS for 10 min. Cells were then washed three times with PBS, rinsed with distilled water, and permeabilised with acetone at −20°C for 10 min. Cells were washed with PBS and incubated with the diluted anti-hScrib C-terminus and anti-ZO-1 antibodies (ZYMED, San Francisco, CA, USA) to stain the tight junctions, or anti-E-cadherin (BD Trasnduction laboratories, Lexington, KY, USA) to stain the adherens junctions for 30 min at room temperature. Cells were then washed three times with PBS and incubated with rhodamine or FITC-conjugated secondary antibodies (Sigma, St Louis, MO, USA), and then washed three times with PBS. Cells were mounted on a slide glass and examined by confocal fluorescence microscopy (Zeiss LSM 410 microscope). Images were captured with a CCD camera.
Detection and typing of HPV DNA by PCR with consensus primers in the L1 ORF
The presence and type of HPV were determined by a PCR-based assay (L1-PCR), originally described by Yoshikawa et al (1991) and modified by Nakagawa et al (1999). The L1 region was amplified in 40 PCR cycles of 1.5 min at 95°C, 1.5 min at 48°C, and 2 min at 70°C, using consensus L1 primers L1C1 (5′-CGTAAACGTTTTCCCTATTTTTTT-3′, 1 M), L1C2 (5′-TACCCTAAATACTCTGTATTG-3′, 0.5 M), and L1C2M (5′-TACCCTAAATACCCTATATTG-3′, 0.5 M). Each reaction product (10 l) was electrophoresed on a 4% agarose gel, stained with ethidium bromide, and viewed under UV light. HPV types were identified based on restriction fragment length polymorphism (RFLP). The initial typing of amplified HPV DNA was carried out by the double digestion with DdeI and RsaI, and then confirmed by digestion with at least three enzymes selected from AccI, AluI, BstXI, FokI, HaeIII, HinfI, KpnI, MaeI, MaeIII, PstI, and XbaI. This L1-PCR can type at least 25 genital HPVs (type 6, 11, 16, 18, 30, 31, 33–35, 42–45, 51–56, 58, 59, 61, 66, 68, and 70).
Immunohistochemistry and immunofluorescence of uterine cervical tissues
Normal cervical tissues and cervical neoplastic tissues were taken in pairs from patients with cervical cancer or its precursor lesions who gave written informed consent. The cervical tissues, which were used in this study, are as follows: normal cervix: 12 cases, cervical condyloma: five cases, low-grade squamous intraepithelial lesions (L-SIL): six cases, high-grade squamous intraepithelial lesions (H-SIL): 12 cases, and invasive cervical cancers: 16 cases. All the condylomas and the cervical neoplasm were positive for low-risk HPV 6/11 and high-risk HPV 16/18, respectively, except one HPV-negative case with the H-SIL. The tissue blocks were embedded in Tissue-Tek OCT compound and quickly frozen in dry ice acetone. Frozen sections (4 μm) were taken on silane-coated glass slides. Sections were fixed in cold acetone for 10 min at −20°C and then rinsed in PBS. Slides were incubated with 10% normal pig serum for 20 min at room temperature to block nonspecific staining and then incubated with the anti-hScrib C-terminus antibody for 60 min at 37°C. Slides were rinsed in PBS and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H+L chain) (Medical and Biological Laboratories, Nagoya, Japan) as the second antibody for 60 min at 37°C. After washing in PBS, the sites of HRP were visualised by 3,3′-diaminobenzidine tetrahydrochloride and 30% hydrogen peroxide in PBS at room temperature. Slides were washed in distilled water and counterstained with 5% methyl green for 15 min. Slides were washed in distilled water and dehydrated through alcohol and xylene, and then mounted with a coverslip.
The immunohistochemical staining patterns of hScrib were compared between the normal epithelium and cervical neoplasm. We also analysed the precise localisation of hScrib in normal endocervical tissues in connection with the junctional proteins by immunofluorescence.
Quantitative RT–PCR analysis of the hScrib mRNA
Total RNA was extracted from all the tumour specimens and the corresponding normal cervical tissues using the RNeasy Mini Kit (Qiagen, Inc., Chartsworth, CA, USA). From 20 μg of total RNA, mRNA was isolated by the Oligotex-dT Super (TaKaRa Biomedicals, Japan). For the RT–PCR, the total volume (10 μl) of mRNA preheated at 65°C for 5 min served as a template for single-strand cDNA synthesis in a 20 μl reaction mixture containing 3 mM MgCl2, 75 mM KCl, 50 mM Tris-HCl (pH 8.3), 0.5 mM dNTPs, 200 μ M oligo (dT) primer, 20 U of RNase inhibitor, and 200 U of M-MLV reverse transcriptase (Gibco BRL, Gaithersburg, MD, USA) at 37°C for 60 min. The reaction was terminated at 95°C for 3 min. To ensure the fidelity of mRNA extraction and RT reaction, all samples was subjected to PCR amplification with oligonucleotide primers specific for the constitutively expressed housekeeping gene GAPDH. At least three different PCR reactions were performed using the LightCycler-FastStart DNA Master SYBR Green I kit (Roche) for each sample. The expression level of hScrib mRNA was standardised as a ratio to the level of GAPDH mRNA. The primer sequences for hScrib were as follows: forward, 5′-GGSTCCATGCTCAAGTGCATCCCGC-3′ and reverse, 5′-GCGGCCGCTTAGAAGTTGGCCACCT-3′. PCR amplifying condition started at 95°C for 10 min, followed by 37 cycles of 95°C for 15 s, 67°C for 10 s, and 72°C for 18 s.
Results
Construction of the anti-hScrib antibody
To generate anti-hScrib antibody, a synthetic peptide sequence of the C-terminus (amino acids 1208–1632) of hScrib was selected (Figure 1A). With the prepared anti-hScrib C-terminus antibody, we analysed the extracts of 293 T cells by Western blot analysis to test whether the anti-hScrib antibody can detect the hScrib protein specifically. The anti-hScrib antibody detected a sharp band around 220 kDa, which corresponds to the in vitro-translated hScrib (Figure 1B). In contrast, it did not react to the in vitro-translated human homologue of Drosophila Discs-Large (hDlg), another PDZ domain containing protein. We concluded that the anti-hScrib antibody detects the hScrib protein specifically.
Construction of the anti-hScrib antibody using its C-terminus. (A) Structure of hScrib consists of 16 canonical LRRs, followed by LAPSD (LAP-specific domain) and four PDZ domains. We generated an anti-hScrib antibody (anti-hScrib C-terminus) using C-terminus (AA 1208–1630) as antigens. (B) Anti-hScrib C-terminus antibody detected the endogenous hScrib protein with 220 kDa in molecular weight in 293 T cells (lane 1) and in vitro-translated hScrib (lane 2), but not in vitro-translated hDlg by Western blotting (lane 3).
hScrib localises at the basolateral membrane in the epithelial cell line
Drosophila scribble localises at the septate junction, the equivalent of the mammalian tight junction. To test whether the endogenous hScrib localises at the basolateral membrane, especially at the tight junction, we analysed its localisation in the polarised canine epithelial cell line MDCK. Looking at the cross-section of the fluorescence image under a confocal microscope, the endogenous hScrib localised at the broad basolateral membrane (Figure 2A and B). These data indicate the conserved homology of amino-acid sequence of scribble among the species. HScrib colocalised with the adherens junctional protein, E-cadherin, at most parts of the basolateral wall excluding the top of the adherens junction (Figure 2A), but not with ZO-1 (Figure 2B). HScrib localised just below the tight junction.
Confocal images of immunofluorescence staining of hScrib, E-cadherin, and ZO-1 in the MDCK epithelial cell line. (A) Partial colocalisation of hScrib and E-cadherin at the basolateral membrane. (B) Localisation of hScrib and ZO-1. ZO-1 localised at the top of the basolateral membrane, where hScrib did not localise.
hScrib is a basolateral membrane-associated protein in primary human tissues
To examine whether hScrib shows the basolateral localisation in primary human tissue, as it showed in the epithelial cell line MDCK, we analysed its localisation in uterine cervical tissues. HScrib localised at the cell–cell boundaries in the normal cervical epithelia. In the columnar endocervical tissues, the immunofluorescence analysis revealed the typically basolateral localisation of hScrib (Figure 3A) and the colocalisation between hScrib and E-cadherin (Figure 3B), which is identical to the observation in cultured MDCK cells (Figure 2A).
hScrib shows a dramatic decrease in the expression during the progression of disease
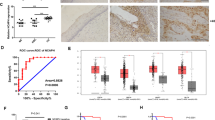
To determine if hScrib is involved in the cervical carcinogenesis, we analysed its expression and localisation in cervical cancer and its precursor lesions (Figure 4). The expression and localisation of hScrib in the cases of L-SIL was indistinguishable from that in the normal cervical epithelium (Figure 4A and D). In contrast to the L-SIL, hScrib showed a dramatic decrease in its expression in the cases of H-SIL (Figure 4E). There was a faint expression of hScrib only in the upper layer of the epithelia of H-SIL (Figure 4E). In contrast to hScrib, the expression of the adherens junction protein E-cadherin was retained along the basolateral membrane in the same case (data not shown). In the normal counterpart tissues, normal expression and localisation of hScrib was confirmed (Figure 4A and B). Human scribble showed the cell–cell boundary-associated localisation in the cases of cervical condyloma, which is a low-risk HPV-related wart (Figure 4C). In the invasive cervical cancer tissues, hScrib showed misdistributed localisation and fully decreased expression comparing its localisation and expression in the normal counterpart tissue of the case (Figure 4F). The other PDZ-domain protein hDlg, which is the substrate of HPV E6 for ubiquitin-mediated degradation, also showed a dramatic decrease in its expression with progression of disease (SN and ST, unpublished data), as described by Watson et al (2002). We compared the expression level of hScrib protein by Western blotting between cervical neoplasm and normal cervical tissues to see whether hScrib actually shows a decrease in the expression compared with the normal counterparts during the progression of disease. Western blotting could not detect any difference between the neoplasm and its normal counterpart in the expression of hScrib in the cases of L-SIL (Figure 5, lanes 1 and 2). In contrast, it showed a clear difference between the neoplasm and its counterpart in the cases of H-SIL and invasive cancer (Figure 5, lanes 3–6), confirming our data of immunohistochemistry. Although hScrib showed a dramatic decrease in its expression with disease progression in the HPV-positive cervical neoplasms, its expression was retained throughout the epithelial layer of the HPV-negative H-SIL case (Figure 6). Next we examined whether the decreased expression of hScrib is due to the reduced transcription of hScrib gene. The mean hScrib/GAPDH mRNA level analysed by the quantitative RT–PCR in all cervical neoplastic tissues and normal control tissues did not show a significant difference in normal cervical tissue (1.40±0.15), L-SIL (1.42±0.21), and H-SIL (1.33±0.08), except for the invasive cancer (0.31±0.16). These data suggest the possibility that degradation of hScrib by E6 and E6AP could have a role in the progressive decrease of hScrib expression during the disease progression from L-SIL to H-SIL.
Immunohistochemical detection of hScrib in normal uterine cervical epithelia and neoplasm. There was a dramatic decrease in the expression of hScrib with disease progression. (A) Normal squamous epithelia. (B) Endocervical epithelia. (C) Cervical condyloma, an HPV-related wart. (D) L-SIL (CIN1). (E) H-SIL (CIN3). (F) Invasive cervical cancer.
Western blot analysis of hScrib protein levels in cervical neoplasm and the normal counterparts using the anti-hScrib C-terminus antibody. (A) The representative result of three independent experiments. (B) Analysis of hScrib protein expression relative to α-tubulin in the cervical neoplasm. The results are expressed as the mean percentage of the normal counterpart. Lanes 1, 3, and 5: the normal counterpart; Lane 2: L-SIL; Lane 4: H-SIL; Lane 6: invasive cancer.
Discussion
Drosophila scribble was identified as an apical-basolateral polarity determinant in the epithelia (Bilder and Perrimon, 2000). Loss of scribble mutation leads to the overgrowth of follicular epithelia and imaginal disc in Drosophila (Bilder et al, 2000b). Loss of polarity and tissue architecture is a common feature of cancer tissues. We sought to observe whether the human homologue of scribble is involved in human carcinogenesis. We showed that hScrib is a substrate of high-risk HPVs and E6AP for ubiquitin-mediated degradation (Nakagawa and Huibregtse, 2000). These data let us to analyse the involvement of hScrib in the uterine cervical carcinogenesis. The multistage nature of cervical carcinogenesis is suitable to investigate its involvement in cancer development. Before addressing this issue, we analysed its localisation in cultured epithelial cells and cervical tissues to see whether hScrib shows the basolateral localisation. Human scribble localised at the broad basolateral membrane in the epithelial MDCK cell line and human endocervical tissues as expected. Its localisation is not restricted to the narrow tight junction, and it colocalised rather with E-cadherin, the adherens junction protein. Drosophila scribble shows the colocalisation with the septate junction marker Coracle and does not colocalise with the adherens junction marker Armadillo, the homologue of vertebrate β-catenin (Bilder et al, 2000b; Bilder and Perrimon, 2000). The arthropod septate junction is thought to be functionally, but not structurally, analogous to the vertebrate tight junction. Drosophila scribble and hScrib have the conserved amino-acid sequence, especially in LRRs and PDZ domains, which propose an identical function for these two homologues. Another PDZ-domain protein, Drosophila Discs-Large, localises at the septate junction, but its human homologue hDlg localises at the basolateral membrane, and not just at the tight junction (Ide et al, 1999). These data are in line with our data with respect to the localisations of scribble at the septate junction in Drosophila embryonic epithelia and those of hScrib at the broad basolateral wall in the mammalian epithelial cells. Recently, we showed that the green fluorescent protein (GFP)-hScrib colocalised with ZO-1, the component of the tight junction in MDCK cells (Nakagawa and Huibregtse, 2000). There can be several reasons why the endogenous localisation of hScrib is different from that of the GFP-hScrib. GFP could misdistribute its original localisation. It is also likely that the vertebrate junctions consist of the completely assembled protein complex and the exogenous protein could not join it.
The normal squamous epithelium consists of multilayers of epithelial cells. In the present study, hScrib showed the localisation at the extensive cell–cell boundaries in the squamous epithelium, in contrast to its basolateral localisation in the endocervical columnar epithelium. The multistage nature of the cervical carninogenesis enabled us to investigate whether hScrib is involved in this process (de Boer et al, 1999). The localisation of hScrib at the cell–cell boundaries and its expression in L-SILs, in which the upper two-third of the epithelium has undergone cytoplasmic differentiation and cells in the lower one-third lack normal differentiation and maturation, was indistinguishable from that in the normal squamous epithelium. In contrast, the expression of hScrib diminished with the progression of disease from L-SILs to invasive cancers. In H-SILs, which was determined by the abnormal changes involving more than lower two-thirds of the epithelium, the expression of hScrib was almost negative, only leaving a very faint expression in the uppermost layer. In invasive cancers, immunohistochemistry could detect the very weak signals of hScrib in the cytoplasm arbitrarily and no hScrib expression at the cell–cell boundaries, which reflects the disruption of cell–cell junctions and mislocalisation of junctional proteins in invasive cancers. Our data show the evidence of involvement of hScrib in the human cancer development for the first time. The dramatic change in the expression of hScrib suggests that its function as a tumour suppressor could be disrupted during the process of progression from L-SIL to H-SIL. No significant difference in the level of hScrib mRNA expression between L-SIL and H-SIL suggests the possibility that the progressive decrease in the expression of hScrib during the development of cervical cancer especially from L-SIL to H-SIL is caused by degradation depending on HPV E6 and E6AP. The retention of the expression of hScrib in the HPV-negative H-SIL case also supports this possibility (Figure 6). To confirm the involvement of HPV E6 and E6AP more precisely, we need to analyse more HPV-negative H-SIL cases and invasive cancers. A cooperative role of downregulation of hScrib mRNA expression and ubiquitin-mediated degradation of hScrib by E6 and E6AP might lead to the complete decrease of hScrib expression during the process of carcinogenesis from H-SIL to invasive cancer.
Our data indicate that hScrib is involved in human carcinogenesis. Most of the mutations in about 50 tumour suppressor genes identified in Drosophila give rise to only overproliferation. Only three genes, scribble, Dlg, and Lethal giant larvae (Lgl), are responsible for retaining tissue organisation (Greaves, 2000; Wodarz, 2000; Bilder et al, 2000b). Mutation of these three genes shares the phenotype of loss of tissue architecture and overgrowth in embryonic imaginal disc and follicular epithelia (Bilder et al, 2000b). These three genes are grouped together as malignant neoplastic tumour suppressors and act in concert to regulate cell proliferation and tissue organisation. The two PDZ-domain proteins, scribble and Dlg, are thought to bind transmembrane protein and form a protein complex as the scaffold at the septate junction. In contrast to the PDZ-domain proteins, Lgl participates in the determination of cell polarity by targeted secretion of membrane proteins. The yeast homologue of Lgl binds to t-SNARE proteins, which is essential for the cargo-carrying vesicle targeting (Lehman et al, 1999; Msch et al, 2002). These three tumour suppressors have the individual human homologue, hScrib, hDlg, and human homologue of Drosophila tumour suppressor lethal giant larvae. High-risk HPVs target hScrib and hDlg for ubiquitin-mediated degradation depending on the E6AP ubiquitin-protein ligase (Nakagawa and Huibregtse, 2000) (For hDlg, unpublished data of SN and JMH). E6 and E6AP could degrade these two scaffolding PDZ tumour suppressors at the basolateral membrane and disrupt the junctional protein assembly at the basolateral region.
Our data indicate the involvement of hScrib in the development of human cancer and shed light on the closely related mechanism between construction of tissue architecture and prevention of cancer development. Further investigation of the interaction among these three tumour suppressors in epithelial cells and cancer cells will give us more knowledge regarding carcinogenesis. These studies are warranted and ongoing in our laboratory.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Bilder D, Birnbaum D, Borg JP, Bryant P, Huigbretse J, Jansen E, Kennedy MB, Labouesse M, Legouis R, Mechler B, Perrimon N, Petit M, Sinha P (2000a) Collective nomenclature for LAP proteins. Nat Cell Biol 2: E114
Bilder D, Li M, Perrimon N (2000b) Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289: 113–116
Bilder D, Perrimon N (2000) Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403: 676–680
de Boer CJ, van Dorst E, van Krieken H, Jansen-van Rhijn CM, Warnaar SO, Fleuren GJ, Litvinov SV (1999) Changing roles of cadherins and catenins during progression of squamous intraepithelial lesions in the uterine cervix. Am J Pathol 155: 505–515
Greaves S (2000) Growth and polarity: the case for scribble. Nat Cell Biol 2: E140
Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA 92: 5249
Huibregtse JM, Scheffner M, Howley PM (1993) Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol 13: 775–784
Ide N, Hata Y, Nishioka H, Hirao K, Yao I, Deguchi M, Mizoguchi A, Nishimori H, Tokino T, Nakamura Y, Takai Y (1999) Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene 18: 7810–7815
Jou TS, Nelson WJ (1998) Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J Cell Biol 142: 85–100
Kaibuchi K, Kuroda S, Fukata M, Nakagawa M (1999) Regulation of cadherin-mediated cell–cell adhesion by the Rho family GTPases. Curr Opin Cell Biol 11: 591–596
Kiyono T, Hirakawa A, Fujita M, Hayashi Y, Akiyama T, Ishihashi M (1997) Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the prosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA 94: 11612–11616
Kuerbitz SJ, Plunkett BS, Walsh WV, Kastan MB (1992) Wild-type p53 is a cell cycle checkpoint determinant following irradiation. Proc Natl Acad Sci USA 89: 7491–7495
Lehman K, Rossi G, Adamo JE, Brennwald P (1999) Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol 146: 125–140
Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362: 847–849
Mantovani F, Massimi P, Banks L (2001) Protrosome-mediated regulation of the hDlg tumor suppressor protein. J Cell Sci 114: 4285–4292
Msch A, Cohen D, Yeaman C, Nelson WJ, Rodriguez-Boulan E, Brennwald PJ (2002) Mammalian homolog of Drosophila tumor suppressor lethal (2) giant larvae interacts with basolateral exocytic machinery in madin-darby canine kidney cells. Mol Biol Cell 13: 158–168
Nakagawa S, Yoshikawa H, Kimura M, Kawana K, Matsumoto K, Onda T, Kino N, Yamada M, Yasugi T, Taketani Y (1999) A possible involvement of aberrant expression of the FHIT gene in the carcinogenesis of squamous cell carcinoma of the uterine cervix. Br J Cancer 79: 589–594
Nakagawa S, Huibregtse JM (2000) Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol 20: 8244–8253
Scheffner M, Huibregtse JM, Howley PM (1994) Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci USA 91: 8797–8801
Scheffner M, Huibregtse JM, Vierstra RD, Howley PM (1993) The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75: 495–505
Watson RA, Rollason TP, Reynolds GM, Murray PG, Banks L, Roberts S (2002) Changes in expression of the human homologue of the Drosophila discs large tumour suppressor protein in high-grade premalignant cervical neoplasias. Carcinogenesis 23: 1791–1796
Wodarz A (2000) Tumor suppressors: linking cell polarity and growth control. Curr Biol 10: R624–R626
Yoshikawa H, Kawana T, Kitagawa K, Mizuno M, Yoshikura H, Iwamoto A (1991) Detection and typing of multiple genital human papillomaviruses by DNA amplification with consensus primers. Jpn J Cancer Res 82: 524–531
Acknowledgements
We thank T Kanda and H Kawakami for their scientific and technical support. This study was supported by the Kanzawa Medical Research Foundation and Grant-in-aid (Number 14571541, SN) for Scientific Research from the Ministry of Education, Science and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Nakagawa, S., Yano, T., Nakagawa, K. et al. Analysis of the expression and localisation of a LAP protein, human scribble, in the normal and neoplastic epithelium of uterine cervix. Br J Cancer 90, 194–199 (2004). https://doi.org/10.1038/sj.bjc.6601465
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6601465
Keywords
This article is cited by
-
Myosin Vb as a tumor suppressor gene in intestinal cancer
Oncogene (2022)
-
The Scribble family in cancer: twentieth anniversary
Oncogene (2020)
-
The Hippo component YAP localizes in the nucleus of human papilloma virus positive oropharyngeal squamous cell carcinoma
Journal of Otolaryngology - Head & Neck Surgery (2017)
-
Loss of the cell polarity determinant human Discs-large is a novel molecular marker of nodal involvement and poor prognosis in endometrial cancer
British Journal of Cancer (2016)
-
HPV-16 impairs the subcellular distribution and levels of expression of protein phosphatase 1γ in cervical malignancy
BMC Cancer (2015)