Abstract
Different therapeutic strategies are currently evaluated in spinal muscular atrophy (SMA) that are aimed at increasing full-length (FL) mRNA levels produced from the SMN2 gene. Assays measuring SMN mRNA levels are needed. We have developed a sensitive, comparative assay based on multiplex fluorescent reverse-transcription polymerase chain reaction (RT–PCR) that can measure, in the same reaction, the levels of SMN mRNA with and without exon 7 sequences as well as those of total SMN mRNA. This assay allows to calculate directly the ratios of FL SMN mRNA to SMN mRNA without exon 7 (Δ7). We have used this assay to compare the levels of SMN transcripts in the blood of 75 unrelated normal subjects and of 48 SMA patients, and in muscle samples of 8 SMA patients. The SMN1 and the SMN2 genes produced very similar levels of total mRNA. Levels of transcripts lacking exon 7 were linearly dependent on the number of SMN2 copies, both in SMA patients and in controls. In patients, FL mRNA levels correlated with SMN2 copy number. A significant but weaker inverse correlation was also observed between FL or Δ7 mRNA levels and disease severity, but patients with three SMN2 copies and different SMA types displayed similar mRNA levels. A significantly higher FL to Δ7 ratio was measured in blood cells than in skeletal muscle (0.80±0.18 versus 0.47±0.11). This assay can be used as a sensitive biomarker for monitoring the effects of various drugs in forthcoming clinical trials of SMA.
Similar content being viewed by others
Introduction
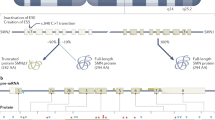
Spinal muscular atrophy (SMA), which is characterized by degeneration of lower motor neurons associated with muscle paralysis and atrophy, represents a frequent recessive autosomal disorder caused by mutation of the survival motor neuron (SMN1) gene at 5q11-q13, consisting mainly of homozygous deletions or conversion events.1 All patients have at least one copy of the highly homologous SMN2 gene, which is located in a more centromeric position on the same chromosome. Both genes code for the SMN protein and differ only at five nucleotide positions, including one within an exonic splicing regulatory element in exon 7.2, 3, 4 Thus, two main mRNA species are produced from the SMN2 gene: a full-length (FL) mRNA and mRNA lacking exon 7 sequences (Δ7), while the SMN1 gene is thought to produce only mRNAs including exon 7.1 Only FL mRNA carries the complete open reading frame required for the production of a stable, active SMN protein. SMA phenotypes range in severity from early death to close to normal life expectancy and have been classified into four groups of decreasing severity: type I (MIM 253300), severe infantile acute SMA, or Werdnig–Hoffmann disease; type II (MIM 253550), or infantile chronic SMA; type III (MIM 253400), juvenile SMA, or Kügelberg–Welander disease; and type IV (MIM 271150) or adult-onset SMA.5
An inverse correlation between SMN2 copy number, SMN protein level and disease severity has been suggested.6, 7 This observation has prompted several groups to examine therapeutic strategies in SMA by increasing FL mRNA levels produced from the SMN2 gene. Cell culture studies have emphasized the potential use of several drugs for this purpose, including sodium valproate and hydroxyurea, which increase FL SMN mRNA either by increasing total SMN mRNA levels or by modulating splicing and facilitating inclusion of exon 7.8, 9, 10, 11, 12, 13, 14 Alternative approaches to aiming at producing higher levels of FL mRNA from the SMN2 gene are based on various forms of antisense oligonucleotides or on bifunctional synthetic RNA-peptide molecules capable of shifting the splicing machinery towards inclusion of exon 7.15, 16, 17 As all these strategies are based on modulation of SMN expression at the transcriptional or at the RNA processing level, clinical trials that are envisaged for the somatic correction of SMA require sensitive and reliable assays, capable of monitoring the effects of the treatments on SMN mRNA levels.18
We describe here a new assay for measuring SMN mRNA levels and compare the expression of the SMN2 gene in peripheral blood cells and in the skeletal muscle of SMA patients. This assay is inspired by the QMPSF method (quantitative multiplex PCR of short fluorescent fragments) that was developed for comparative quantitative analysis of genomic DNA,19, 20 but is applied here to reverse-transcribed mRNA.
Subjects and methods
Subjects
We collected blood samples from 75 unrelated control subjects and 48 SMA patients who did not receive any treatment. Patients fulfilled the diagnostic criteria of proximal SMA defined by the International SMA Consortium.5 Fourteen patients with severe, generalized muscle weakness at birth or within the first 6 months were classified as type I SMA, 22 patients who were able to sit although they could not walk unaided were classified as type II and 12 patients who were able to walk before the onset of proximal muscle weakness were classified as type III. Blood was collected after overnight fasting into PAXgene Blood RNA Tubes (QIAGEN®, Courtabœuf, France) for RNA extraction, into ethylenediamine tetraacetic acid (EDTA) tubes for SMN genotyping and into BD Vacutainer® CPT cell preparation tube with lithium heparin for protein isolation; samples were treated after transport at room temperature for times ranging from a minimum of 2 h to a maximum of 3 days. Quantification of SMN1 and SMN2 copies was performed using a multiplex PCR assay.19 SMA patients had homozygous deletions or conversions of the SMN1 gene and carried at least two copies of the SMN2 gene.
Striated paravertebral muscles were taken as surgery residues during cervical arthrodesis of eight SMA patients. Muscle samples were immediately frozen in liquid nitrogen and stored at −80°C. A section of each sample (∼10 mg) was used for RNA extraction. Blood was collected from the same patients as described above.
Blood or muscle samples were obtained after informed and written consent from each participant or the legal guardian according to a protocol reviewed and approved by the Local Ethics Committee.
Reverse-transcription polymerase chain reaction of SMN transcripts
Total cellular RNA was extracted from whole blood using the PAXgene Blood RNA kit (Qiagen) and from muscles RNA using Tripure reagent (Roche Diagnostics, Indianapolis, IN, USA). Total RNA quality and quantity were evaluated for each RNA sample using the RNA 6000 Nano Assay (Agilent Technologies, Massy, France) after RNAse-free DNAse treatment (Roche Diagnostics). One hundred nanograms of total RNA was reverse-transcribed into complementary DNA using the Sensiscript Reverse Transcription kit (Qiagen) in the presence of Recombinant RNasin (Promega, Lyon, France). First-strand synthesis from total RNA was carried out using an oligo d(T)18 mRNA Primer (New England Biolabs, Saint Quentin en Yvelines, France).
The single-stranded cDNA was PCR-amplified using two pairs of primers to cover different SMN mRNA regions and with two additional primer pairs that amplify reference mRNAs (Table 1). Sense primers were 5′-labelled with 6-FAM. All primers were used in a single PCR of 25 μl. PCRs were carried out using 2 mM MgCl2, 1.5 unit of Thermoprime plus DNA polymerase (ABgene, Epson, UK), 0.4 μ M of each primer and 4 μl of cDNA. After an initial cycle of denaturation at 94°C for 4 min, 24 cycles were performed consisting of denaturation at 94°C for 10 s, annealing at 58°C for 15 s and extension at 72°C for 20 s, and final extension at 72°C, in a PCR System 9700 (Applied Biosystems, Courtabœuf, France). Quantitative analyses were carried out using the GeneScan 3.7 software (Applied Biosystems) after electrophoretic separation of the reverse-transcription polymerase chain reaction (RT–PCR) products on a DNA sequencer (ABI 3100; Applied Biosystems).
Data calculation and statistical analysis
The values obtained for the different mRNA forms were normalized, using the mean of peak heights, by calculating the ratios of SMN Δ7 mRNA to MLH1 mRNA (Δ7/MLH1) or to HPRT mRNA (Δ7/HPRT), and of SMN FL mRNA to MLH1 mRNA (FL/MLH1) or to HPRT mRNA (FL/HPRT). Global levels of SMN transcripts were normalized by calculating the ratios of exon 1–2b mRNA to MLH1 mRNA (total/MLH1) or exon 1–2b mRNA to HPRT mRNA (total/HPRT). The relative contribution of exon 7 splicing was evaluated by calculating the ratio of full-length to Δ7 mRNA (FL/Δ7). Each measurement was realized in triplicate and all data are expressed as mean±standard deviation (SD).
Correlations between SMN mRNA levels and gene copy numbers or between SMN mRNA levels and SMA type were tested using the nonparametric Kruskal–Wallis test combined and the Mann–Whitney U-test with Bonferroni correction. For all tests, P<0.05 (or P<0.017 after Bonferroni adjustment) were considered statistically significant. All statistical analyses were conducted using MedCalc® software (Version 8.2, Mariakerke, Belgium).
Results
Multiplex fluorescent RT–PCR assay
Our goal was to develop a new SMN mRNA assay aiming at measuring simultaneously the variation of total SMN transcripts, of FL SMN mRNA and of Δ7 SMN mRNA. As shown in Figure 1a, we choose to develop a fluorescent multiplex RT–PCR assay able to amplify in the same reaction total SMN transcripts using primers in exons 1 and 2b (P1 and P2, respectively), FL and Δ7 SMN transcripts using primers in exons 6 and 8 (P3 and P4, respectively) as well as HRPT and MLH1 mRNA transcripts used as internal controls. We tested several candidate housekeeping gene mRNAs (ACTB, HMBS, MLH1, HPRT) on total RNA samples from 10 individuals after precise dosage using the RNA 6000 Nano Assay from Agilent Technologies. We found that the HPRT and the MLH1 mRNAs were the least variable since these internal references yielded similar and consistent results on peripheral blood total RNA samples, with standard variations of 25 and 27%, respectively, measured over 10 control subjects with the same SMN genotype (data not shown). A typical profile, obtained from a control individual with two copies of the SMN1 gene and two copies of the SMN2 gene, is shown in Figure 1b. Very similar results were obtained using the MLH1 or the HPRT value for normalization. Thus, in the following, we will present data normalized using the HPRT mRNA reference.
Semi-quantitative fluorescent RT–PCR. (a) Patient and control SMN cDNAs were PCR-amplified using two pairs of primers: P1–P2, yielding a 286 bp product that measures total SMN mRNA and P3–P4, which yielded fragments of 184 and 130 bp from full-length (FL) and Δ7 cDNA, respectively. MLH1 and HPRT cDNAs were amplified as internal references. (b) RT–PCR products were loaded on an ABI Prism 3100 DNA sequencer and profiles were analyzed using the GeneScan 3.7 Software. The diagram shown was obtained from a 2C2T control (two copies of SMN2 and two copies of SMN1). The x axis indicates the fragment size and the y axis the fluorescence in arbitrary units. For each PCR product, peak height was used to calculate the ratios presented in the following figures. Note that the MLH1 and HPRT peaks are used only for normalization and do not necessarily reflect the relative abundance of their mRNA relative to SMN mRNAs. Δ7, without exon 7; FL, full-length; RT–PCR, reverse-transcription polymerase chain reaction; SMN, survival motor neuron.
One should note that this method does not determine the absolute ratios of SMN mRNA species versus MLH1 or HPRT mRNA, because experimental variables affect the signals measured for each amplicon, as discussed below. Normalization of the heights of fluorescence profiles corrects for variations in input RNA and allows comparisons of SMN mRNA species between different samples. To select a cycle number ensuring amplification within the exponential range for each mRNA species of the multiplex RT–PCR, we carried out experiments with different cycle numbers and found that each of the peaks illustrated in Figure 1b doubled between 24 and 25 cycles (data not shown). Although we choose 24 cycles for our routine analyses, one should note that, even using the same PCR conditions as described in Subjects and methods, the optimal cycle number must be determined when setting up the assay with new reagents, because the intensity of each peak reflects not only the amount of the corresponding mRNA species, but also the exact concentration of the primer pairs used for each amplicon and the variable intensities of fluorescent labels. It is therefore essential to use exactly the same PCR conditions and primer dilutions within each series of measurements. Profiles such as the one shown in Figure 1b are then reproducible between identical RNA samples and, for each amplicon, peak values from different samples can be compared reliably after normalization. For example, to evaluate the robustness of the assay, we repeated independently the multiplex PCR on the same control cDNA or we repeated the entire assay, including the reverse transcription step, on the same control RNA, and found that the standard deviation of the multiplex RT–PCR was within 10% (see Supplementary Tables 1 and 2). We have also tested the variation of SMN mRNA levels of a single control individual over 3 consecutive days and at four different times of the day and found that these levels were always within the experimental variation of the method (data not shown).
FL and Δ7 mRNA levels relative to SMN2 copy number and to SMA disease severity
We recruited 48 SMA patients (14 type I, 22 type II and 12 type III) and determined their SMN genotype as shown in Table 2. We then measured their peripheral blood SMN mRNA levels, normalized to HPRT mRNA and expressed them as a function of SMN2 copies. As shown in Figure 2 and Supplementary Table 3, FL, Δ7 and total SMN2 mRNA levels increased linearly with SMN2 copy number. Each of these values was found doubled between 2 and 4 SMN2 copies, and the FL/Δ7 ratios remained close to 0.9. These data demonstrate a tight correlation of SMN2 transcript levels with SMN2 copy number in peripheral blood cell. Even if peripheral blood cells are not the target cells in SMA disease, these data are consistent with the inverse correlation found between SMN2 copy number and disease severity.7 We therefore compared SMN transcript levels to the type of SMA and found that each form of SMN2 transcripts (FL mRNA, Δ7 mRNA and total SMN mRNA) indeed was inversely correlated with disease severity, while the FL/Δ7 ratios remained close to 0.9 (Figure 3 and Supplementary Table 4). The inverse correlation observed between transcript levels and disease severity was, however, weaker than the direct correlation observed with SMN2 copy number. In fact, as shown in Table 2, the three phenotypic groups, and particularly the type III group, are heterogeneous with regard to SMN2 copy numbers. Conversely, one finds in Table 2 that patients with three SMN2 copies are heterogeneous with regard to disease phenotype. We have therefore examined the mRNA levels for each of the three subgroups with three SMN2 copies and found that Δ7 mRNA and FL mRNA levels did not differ significantly that is they appear to be determined by the SMN2 copy number (Figure 4). These data therefore indicate that, in spite of the heterogeneity mentioned above, the inverse correlation observed between mRNA levels and disease phenotype reflects the distribution of genotypes (Table 2) and the strong correlation between mRNA levels and SMN2 copy numbers.
Correlation between SMN2 copy number and levels of SMN transcripts in patient groups. Forty-eight SMA patients were studied: 10 with two copies of SMN2 (2C), 33 with three copies (3C) and 5 with four copies (4C). SMN transcript levels from patient peripheral blood cells are shown normalized to HPRT expression levels. For details, see Supplementary Table 3 (patients). (a), (b) and (c) represent FL SMN mRNA, Δ7 SMN mRNA or total SMN mRNA, respectively. In each panel, vertical bars represent the average of the peak heights of the corresponding amplicon normalized by the peak heights of the HPRT amplicon. (d) FL/Δ7 ratios are shown. Each RNA sample was measured in triplicate. For each group, the numbers shown inside the bar represents the increase in expression level relative to the group with two copies of SMN2. Error bars represent the standard deviation (SD). For global comparisons between the three groups, the Kruskal–Wallis test was used and found significant (P<0.001). The Mann–Whitney test with Bonferroni correction was used to compare expression between groups (*P<0.017, **P<0.0017). Δ7 SMN mRNA, SMN mRNA without exon 7; FL SMN mRNA, full-length SMN mRNA; SMA, spinal muscular atrophy; SMN, survival motor neuron.
Comparison between SMA type and SMN transcript levels in patient groups. The same 48 SMA patients (14 type I SMA, 22 type II SMA and 12 type III SMA, see Table 2) were grouped according to their disease phenotype. For details, see Supplementary Table 4. (a), (b) and (c) The averaged relative values of FL, Δ7 and total SMN transcript, respectively, normalized to the HPRT are shown. (d) shows FL/Δ7 ratios. Statistical analysis was performed as in Figure 2. Δ7, without exon 7; FL, full-length; SMA, spinal muscular atrophy; SMN, survival motor neuron.
Patients with three SMN2 copies display similar mRNA levels, regardless of their SMA type. The subgroups with three SMN2 copies in the second column of Table 2 are compared for the HPRT-normalized expression levels of Δ7 mRNA (a) and FL mRNA (b). Δ7 mRNA, mRNA without exon 7; FL mRNA, full-length mRNA; SMN, survival motor neuron.
Contribution of SMN2 copy number to FL and Δ7 mRNA levels in control individuals
We then genotyped 75 normal control subjects and determined the various forms of SMN transcripts in their peripheral blood cells. To determine the relative contribution of SMN1 and SMN2 copy numbers to FL mRNA levels, we considered only subjects with two copies of the SMN1 gene, that is, 66 of the 75 normal control subjects and measured the different forms of SMN transcripts, for 0, 1 and 2 copies of the SMN2 gene (Figure 5 and Supplementary Table 3). In Figure 5, panel a shows that FL mRNA levels correlate with the number of SMN2 copies present in addition to the two SMN1 copies. Panel b shows that Δ7 mRNA levels double from one to two SMN2 copies. It is worth noting that, in the six control individuals lacking the SMN2 gene, no Δ7 transcript was detected.
Comparison of SMN copy numbers and SMN transcript levels in controls. SMN transcript levels were measured in 66 control individuals with constant number of SMN1 (two copies, shown as 2T) and no SMN2 (0C2T, six individuals) or one copy of SMN2 (1C2T, 30 individuals) or two copies of SMN2 (2C2T, 30 individuals). For details, see Supplementary Table 3 (controls). (a) Normalized and averaged FL SMN mRNA levels. (b) Normalized and averaged Δ7 SMN mRNA levels. Transcript levels were measured in triplicate and were normalized using HPRT mRNA levels. Error bars represent the SD. Note, in (b), that no Δ7 mRNA could be detected in the six individuals with no copy of the SMN2 gene. Statistical analysis was performed as in Figure 2. Δ7 SMN mRNA, SMN mRNA without exon 7; FL SMN mRNA, full-length SMN mRNA; SMN, survival motor neuron.
Similar steady-state levels of total transcripts from the SMN1 and the SMN2 genes
It has been shown that the SMN1 and the SMN2 genes share quasi-identical promoter sequences and that the activities of these promoters, when tested using reporter constructs, are very similar.22, 23 Our assay allowed us to compare the in vivo expression levels of both genes, since it included an amplicon measuring total transcript levels. We therefore compared total transcript levels of patients with two SMN2 copies with those of control subjects with two SMN1 copies and no SMN2 copy, and found that total SMN transcript levels did not differ between the two groups (data not shown). This demonstrates that steady-state levels of total transcripts from the SMN1 and from the SMN2 gene are similar. Therefore, if Δ7 mRNA and FL mRNA have similar stability, the SMN1 and SMN2 promoters must have the same strength in vivo, in keeping with their sequence conservation and the promoter strength measured with reporter constructs.22, 23, 24
Different FL/Δ7 ratio in skeletal muscle and in peripheral blood cells
We then asked the question of the relative proportions of the two main forms of SMN2 transcripts in cell types that are more relevant to SMA. We therefore compared SMN2 transcripts from blood and from muscle samples obtained from one type I, six type II and one type III SMA patients, all with three copies of SMN2. As shown in Figure 6, the FL/Δ7 ratio in skeletal muscle from these patients was 0.47±0.11 and was significantly different from the ratio in peripheral blood cells (0.80±0.18; P<0.005; t-test). The ratios, in muscle, for the single type I patient and for the single type III patient did not differ significantly from those of the other patients, affected with type II SMA. These observations suggest that the lower FL/Δ7 ratio in skeletal muscle may be due to a different regulation of alternative splicing of exon 7. While the FL/Δ7 ratios can be compared without the need of normalization with a housekeeping gene product, variations of FL mRNA levels and of Δ7 mRNA levels between tissues can be evaluated only by using appropriate reference mRNAs. Since no data concerning the stability of expression of the MLH1 or the HPRT mRNA between skeletal muscle and peripheral blood are presently available, we were not able to compare FL mRNA levels, Δ7 mRNA levels and total SMN mRNA levels between these two tissues.
SMN2 transcript levels in blood and muscle. Blood and muscle samples were obtained from eight SMA patients with three copies of the SMN2 gene and transcript levels were measured in duplicate. The average FL/Δ7 ratios with error bars representing SD are shown. Statistical analysis was performed using a t-test (**P<0.005). Δ7 SMN, SMN without exon 7; FL SMN, full-length SMN; SMA, spinal muscular atrophy; SMN, survival motor neuron.
Discussion
SMN is an ubiquitous and essential component of small nuclear ribonucleoprotein assembly.25 One can therefore envisage that while motor neuron cells are more sensitive to limiting levels of SMN protein, therapeutic modulation of SMN expression could be monitored in other cell types, including peripheral blood cells. Several assays for monitoring SMN mRNA levels in peripheral blood cells have been described.26, 27, 28 These assays were all based on real-time PCR and required the use of different primer pairs that target the FL and the Δ7 mRNAs in separate reactions. Therefore, accurate calculations of expression levels relied on the quality of calibration curves and the FL/Δ7 ratio, which is a key parameter to reveal the effects of treatments that modify the splicing pattern of SMN2 exon 7, was calculated indirectly.28 Our assay differs from recent real-time PCR-based assays mainly because it allows more direct comparison of FL, Δ7 and total SMN mRNA levels. In our multiplex fluorescent RT–PCR assay, FL and Δ7 transcripts are simultaneously PCR-amplified using the same primer pair. This design reduces the intersample variability of FL and Δ7 measurements to a minimum and FL/Δ7 ratios can be calculated directly. Even if the functional implications of the Δ7 protein are still debated, the FL/Δ7 ratio, measured at the mRNA level, is a valuable indicator of the effect of treatments that modulate SMN RNA splicing.29, 30 Moreover, we have also included into the same reaction an amplicon that measures total SMN transcripts. Any significant increase of this value should provide a direct indication of the effect of drugs that increase SMN transcription or SMN mRNA stability. In this regard, an additional advantage of this method is the possibility to include multiple internal RNA references in the same reaction, as shown here for the MLH1 and the HPRT mRNAs. As particular treatments may affect the level of the present reference mRNAs, one may have to include additional references for monitoring the effects of particular drugs in clinical trials. Therefore, the reproducibility and the sensitivity of this assay provide the basis for its use in forthcoming clinical assays of SMA.
The analysis of six control individuals lacking the SMN2 gene did not reveal measurable levels of Δ7 mRNA expressed from the SMN1 gene. Similarly, using real-time PCR, analysis of control individuals lacking SMN2 copies yielded a Δ7 transcript amount close to zero.26, 28 We have also shown that, in peripheral blood cells of both SMA patients (Figure 2) and unaffected controls (Figure 5), the levels of Δ7 transcripts are linearly dependent on the number of SMN2 copies. In patients, FL mRNA levels are also linearly dependent on the SMN2 copy number (Figure 2). Our data are consistent with those obtained by Sumner et al,28 concerning the correlation of SMN mRNA levels with copy numbers. The same study also estimated to 50% the fraction of peripheral blood SMN mRNA lacking exon 7, in good agreement with our data, since we show that the FL/Δ7 ratio in peripheral blood cells of patients is close to 0.9 (Supplementary Tables 3 and 4).
We have also shown in patients a weaker inverse correlation between FL SMN mRNA levels and disease severity (Figure 3). Similarly to Sumner et al,28 we find that the FL and Δ7 mRNA levels of type I patients differ significantly from those of other SMA types. In addition, we find an increase of SMN mRNA levels in SMA type III compared to type II, which is however not significant for Δ7 mRNA. The ability to detect a correlation between FL SMN mRNA levels and the type of SMA appears to be influenced by the heterogeneity of genotypes within each phenotypic group. In our series, the distribution of the genotypes was close to that described by Feldkötter in 375 patients, except for the presence of a slightly higher proportion of type I patients with three copies of SMN2 in our series (Table 2). We have considered in particular the patients with three SMN2 copies, who belong to different phenotypic groups, and found that their FL and Δ7 mRNA levels did not differ (Table 2; Figure 4). This indicates that the genotype determines SMN expression at the mRNA level in blood cells, suggesting different determinants for the disease phenotype.
Finally, we have shown that the FL/Δ7 ratio in blood cells and skeletal muscle of patients differs significantly (0.80±0.18 versus 0.47±0.11), indicating that the Δ7 mRNA is the predominant form in skeletal muscle (Figure 6). To the best of our knowledge, this is the first quantitative comparison of the FL/Δ7 ratios in skeletal muscle versus blood cells of patients with the same genotype. Several studies have described the levels of SMN isoforms in different tissues of patients compared to controls.31, 32, 33, 34 Interestingly, when comparing SMN transcripts from different tissues in control fetuses, Soler-Botija et al34 detected the highest amount of FL SMN in control spinal cord. In SMA spinal cord, they showed a FL/Δ7 ratio close to 1, whereas this ratio was close to 0.5 in SMA skeletal muscle, in very good agreement with our data. In addition, Soler-Botija et al34 demonstrated a 70% reduction of the SMN protein in spinal cord of SMA versus control fetuses and a 35% reduction of SMN protein in skeletal muscle of SMA versus control fetuses. Taken together, these data suggest a regulation of exon 7 alternative splicing in a tissue-dependent manner and raise the question of whether the regulation of SMN transcription and splicing is similar in blood cells and motor neurons in response to a given drug. The generation of motor neuron cell lines carrying the SMN2 gene would be very useful to answer this question. At any rate, this assay can be used to determine if drugs known to affect SMN mRNA levels in cell culture models also have an effect in vivo.
This new SMN mRNA assay ensured reproducible quantitative measurements of SMN mRNA species even on RNA from blood samples stored up to 4 days and/or shipped from distant laboratories. This kind of flexibility, which is essential in view of establishing useful and reliable biomarkers for SMA clinical trials, was not observed in assays of SMN protein levels, which seem to be limited by the instability of the SMN protein during storage of the samples.26 Nevertheless, protein assays should also be developed as biomarkers, and a recently developed cell immunoassay applied to peripheral blood cells is promising.28
References
Lefebvre S, Bürglen L, Reboullet S et al: Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–165.
Cartegni L, Krainer AR : Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 2002; 30: 377–384.
Kashima T, Manley JL : A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 2003; 34: 460–463.
Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR : Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet 2006; 78: 63–77.
Zerres K, Davies KE : Workshop report. 59th ENMC International Workshop: Spinal muscular atrophies: recent progress and revised diagnostic criteria. Neuromusc Disord 1999; 9: 272–278.
Lefebvre S, Burlet P, Liu Q et al: Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16: 265–269.
Feldkötter M, Schwarzer V, Wirth R, Wienker TF, Wirth B : Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet 2002; 70: 358–368.
Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H : Treatment of spinal muscular atrophy by sodium butyrate. Proc Natl Acad Sci USA 2001; 98: 9808–9813.
Andreassi C, Jarecki J, Zhou J et al: Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients. Hum Mol Genet 2001; 10: 2841–2849.
Brichta L, Hofmann Y, Hahnen E et al: Valproic acid increases the SMN2 protein level: a well-known drug as a potential therapy for spinal muscular atrophy. Hum Mol Genet 2003; 12: 2481–2489.
Sumner CJ, Huynh TN, Markowitz JA et al: Valproic acid increases SMN levels in spinal muscular atrophy patient cells. Ann Neurol 2003; 54: 647–654.
Andreassi C, Angelozzi C, Tiziano FD et al: Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet 2004; 12: 59–65.
Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH : Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells. Ann Neurol 2005; 58: 194–202.
Jarecki J, Chen X, Bernardino A et al: Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet 2005; 14: 2003–2018.
Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F : Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA 2003; 100: 4114–4119.
Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ : Correction of SMN2 pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther 2005; 12: 1013–1022.
Baughan T, Shababi M, Coady TH, Dickson HM, Tullis GE, Lorson CL : Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol Ther 2006; 14: 54–62.
Sumner CJ : Therapeutics development for spinal muscular atrophy. NeuroRx 2006; 3: 235–245.
Saugier-Veber P, Drouot N, Lefebvre S et al: Detection of heterozygous SMN1 deletions in SMA families using a simple fluorescent multiplex PCR method. J Med Genet 2001; 38: 240–243.
Saugier-Veber P, Goldenberg A, Drouin-Garraud V et al: Simple detection of genomic microdeletions and microduplications using QMPSF in patients with idiopathic mental retardation. Eur J Hum Genet 2006; 14: 1009–1017.
Jong YJ, Chang JG, Lin SP et al: Analysis of the mRNA transcripts of the survival motor neuron (SMN) gene in the tissue of an SMA fetus and the peripheral blood mononuclear cells of normals, carriers and SMA patients. J Neurol Sci 2000; 173: 147–153.
Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J : The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet 1999; 64: 1365–1370.
Monani UR, McPherson JD, Burghes AH : Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT). Biochim Biophys Acta 1999; 1445: 330–336.
Boda B, Mas C, Giudicelli C et al: Survival motor neuron SMN1 and SMN2 gene promoters: identical sequences and differential expression in neurons and non-neuronal cells. Eur J Hum Genet 2004; 12: 729–737.
Pellizzoni L, Yong J, Dreyfuss G : Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002; 298: 1775–1779.
Brichta L, Holker I, Haug K, Klockgether T, Wirth B : In vivo activation of SMN in spinal muscular atrophy carriers and patients treated with valproate. Ann Neurol 2006; 59: 970–975.
Brahe C, Vitali T, Tiziano FD et al: Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients. Eur J Hum Genet 2005; 13: 256–259.
Sumner CJ, Kolb SJ, Harmison GG et al: SMN mRNA and protein levels in peripheral blood: biomarkers for SMA clinical trials. Neurology 2006; 66: 1067–1073.
Kerr DA, Nery JP, Traystman RJ, Chau BN, Hardwick JM : Survival motor neuron protein modulates neuron-specific apoptosis. Proc Natl Acad Sci USA 2000; 97: 13312–13317.
Le TT, Pham LT, Butchbach ME et al: SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 2005; 14: 845–857.
Gennarelli M, Lucarelli M, Capon F et al: Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun 1995; 213: 342–348.
Burlet P, Huber C, Bertrandy S et al: The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum Mol Genet 1998; 7: 1927–1933.
Jong YJ, Chang JG, Lin SP et al: Analysis of the mRNA transcripts of the survival motor neuron (SMN) gene in the tissue of an SMA fetus and the peripheral blood mononuclear cells of normals, carriers and SMA patients. J Neurol Sci 2000; 173: 147–153.
Soler-Botija C, Cusco I, Caselles L, Lopez E, Baiget M, Tizzano EF : Implication of fetal SMN2 expression in type I SMA pathogenesis: protection or pathological gain of function? J Neuropathol Exp Neurol 2005; 64: 215–223.
Acknowledgements
We thank the patients and their families who have contributed to this project and without whom this study would not have been possible. We thank the SMA group of the Association Française contre les Myopathies (AFM), especially M Roussel and A Goussiaume, and the Tissue Bank for Research of AFM, especially S Vasseur. We thank A Goldenberg, C Vanhulle, D Bonneau, A Moerman, C Cances, P Cintas and D Nicolas, for referring patients and J Bénichou for help in statistical methodology. This work was supported by the Association Française contre les Myopathies (AFM). Healthy volunteers were recruited with the support of the CIC – INSERM – Rouen University Hospital after approval by the Local Ethics Committee.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Rights and permissions
About this article
Cite this article
Vezain, M., Saugier-Veber, P., Melki, J. et al. A sensitive assay for measuring SMN mRNA levels in peripheral blood and in muscle samples of patients affected with spinal muscular atrophy. Eur J Hum Genet 15, 1054–1062 (2007). https://doi.org/10.1038/sj.ejhg.5201885
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201885
Keywords
This article is cited by
-
Biomarkers in 5q-associated spinal muscular atrophy—a narrative review
Journal of Neurology (2023)
-
Survival Motor Neuron Protein is Released from Cells in Exosomes: A Potential Biomarker for Spinal Muscular Atrophy
Scientific Reports (2017)
-
Decay in survival motor neuron and plastin 3 levels during differentiation of iPSC-derived human motor neurons
Scientific Reports (2015)
-
Genetic variations of the A13/A14 repeat located within the EGFR3′ untranslated region have no oncogenic effect in patients with colorectal cancer
BMC Cancer (2013)
-
SMN transcript levels in leukocytes of SMA patients determined by absolute real-time PCR
European Journal of Human Genetics (2010)