Abstract
The m.13513G>A transition in the mitochondrial gene encoding the ND5 subunit of respiratory chain complex I, can cause mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS) and has been reported to be a frequent cause of Leigh syndrome (LS). We determined the frequency of the mutation in a cohort of 123 patients with reduced complex I activity in muscle (n=113) or fibroblast (n=10) tissue. We describe a Pyrosequencing™ assay for rapid detection and quantification of the m.13513G>A mutation. Two patients with the mutation were identified; both had LS, optical atrophy and a Wolff–Parkinson–White Syndrome (WPWS)-like cardiac conduction defect. The clinical presentation of the m.13513G>A mutation is discussed. We conclude that the m.13513G>A mutation seems not as frequent as previously suggested and is most likely to be present in patients with Leigh (-like) syndrome combined with a complex I deficiency, optic atrophy and/ or WPWS. In addition, we confirmed that the adjacent m.13514A>G mutation is a rare cause of LS or MELAS since no cases with this transition were found.
Similar content being viewed by others
Introduction
Leigh syndrome (LS) is a neurodegenerative disease usually starting in childhood with typical lesions in the brainstem and basal ganglia. LS can result from mutations in both mitochondrial (mt) and nuclear genes that cause a deficiency of one of the complexes of the mitochondrial respiratory chain, most often complex I.1 Mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS) also results from deficiencies in mitochondrial energy production. In the majority of cases MELAS is caused by the m.3243A>G mutation in the mtDNA, but several other causative mtDNA mutations have been reported.2, 3 The m.13513G>A transition in the mitochondrial gene encoding the ND5 subunit of respiratory chain complex I, has been reported in both Leigh (-like) and MELAS (-like) syndromes. Until now 21 patients with this mutation have been described; seven with MELAS2, 3, 4, 5 and 14 with LS.6, 7, 8, 9, 10 In 16 of them complex I activity was biochemically tested, and this was found to be reduced in 12 cases. Several authors have postulated that the m.13513G>A mutation is a frequent cause of either LS or MELAS and suggested that it should be routinely tested in these patients 3, 4, 5, 6, 9, 11 even in the absence of a biochemical complex I deficiency.9, 11 The objective of the current study was to determine the frequency of the mutation in a large cohort consisting of 123 patients with an established reduction in complex I enzyme activity. In addition, we review the clinical presentation of the m.13513G>A transition and discuss in which patients it is indicated for a routine molecular diagnostic laboratory, to screen for this mutation. Furthermore, we describe a Pyrosequencing™ assay (PSQ) for rapid detection and quantification of the m.13513G>A mutation. As this assay allows to simultaneously test for the presence of the adjacent, more rare m.13514A>G mutation, all patients were checked for this mutation too. The m.13514A>G mutation affects the same codon as the m.13513G>A transition and has been reported in four patients with a MELAS- or LS-like phenotype.4, 10, 11
Materials and methods
Patients
For the present study we selected patients from our records with a complex I deficiency in muscle or fibroblast tissue, either isolated or combined with a deficiency of one or more of the other complexes. We included both isolated and combined deficient subjects since mutations in complex I genes are known to occasionally cause secondary reductions in other complexes; (Budde et al,12 personal observations). All subjects were initially referred to our molecular diagnostic lab because of clinical suspicion for a mitochondrial respiratory chain disease. In the majority of cases, the referring clinician had provided only very limited clinical information. In total, 123 unrelated patients were included of whom 68 had an isolated complex I deficiency in muscle (n=63) or fibroblasts (n=5) whereas 55 had a combined deficiency of complex I plus at least one of the other mitochondrial respiratory chain complexes in muscle (n=50) or fibroblasts (n=5). Biochemical analyses of muscle and cultured fibroblast tissues was performed as described before.13, 14 All patients were tested for the presence of the m.13513G>A and m.13514A>G mutation in the mitochondrial ND5 gene. All selected subjects had tested negative for common pathogenic point mutations at nucleotide positions m.3243, m.8344 and m.8993 and single large mtDNA rearrangements (deletions or duplications).
Molecular analysis
DNA was isolated from tissue obtained through a biopsy of quadriceps muscle or from cultured skin fibroblasts and DNA was extracted from biopsy material using the Gentra Purgene Genomic DNA isolation kit (Minneapolis, MN, USA) according to the specifications of the manufacturer. Presence of the m.13513G>A and m.13514A>G mutations was tested using Pyrosequencing™ technology (Biotage AB, Uppsala, Sweden), which essentially is a real-time mini-sequencing technique particularly appropriate for rapid SNP genotyping (http://www.pyrosequencing.com). Pyrosequencing involves annealing of a sequencing primer to a PCR amplified single-stranded biotinylated DNA template after which the actual sequencing takes place using a Pyrosequencer PSQ96MA platform.15 During the sequencing process several enzymes and specific substrates are used to generate a light signal. Analysis starts as the enzymes and substrates are dispensed into the reaction mixture. Subsequently, nucleotides are dispensed sequentially (one after the other) into the mixture. Light is generated whenever a nucleotide forms a base pair with the complementary base in the DNA template. The amount of light produced is proportional to the number of nucleotides incorporated. If the added nucleotide is not complementary to the next base in the template no light is produced. Pyrosequencing not only allows rapid testing for the presence of a known point mutation at a specific position, but in case of a mitochondrial DNA mutation also for accurate quantification of the level of heteroplasmy.16, 17, 18 PSQ was performed according to the protocol of the manufacturer. PCR amplification of a mtDNA fragment containing the m.13513 and m.13514 position was carried out using the following primers: universal primer (biotinylated) 5′- GCTGCTCCGGTTCATAGATT-3′; forward primer 5′-CTTCAACCTCCCTCACC-3′; reverse primer 5′-AGCGCTGCTCCGGTTCATAGATTGCTCAGGCGTTTGTGTATGA-3′. PCR was carried out in a 50 μl volume containing 0.18 μ M forward primer, 0.02 μ M reverse primer and 0.2 μ M of the biotinylated universal primer. PCR conditions were 92°C for 30 s, 55°C for 30 s, 72°C for 30 s, for a total of 40 cycles. Single-stranded template DNA, in the present assay the reverse strand of the fragment, was obtained by immobilizing biotinylated PCR fragments using streptavidine-coated sepharose beads, which were then passed through a series of wash steps using a 96-pin Vacuum Prep Workstation. The forward primer used for sequencing was 5′-CACAGGTTTCTACTCCAA-3′ (nt. 13494–13511). The sequence to analyze in case of the m.13513G>A mutation was A(G/A)ACCACAT (nt. 13512–13520). The dispensation order that was applied during Pyrosequencing reaction was TAGAGCACAT. One microgram of single-stranded DNA binding (SSB) protein (USB Corporation, Cleveland, OH, USA) was added to each reaction to reduce secondary structure formation of DNA molecules, thereby increasing signal specificity. The current assay was validated by mixing wild-type and mutant templates in different ratios. Subsequently, the level of heteroplasmy of each mixture was determined by PSQ and results were compared with the expected theoretical values. Mutant templates carrying the m.13513G>A mutation necessary for the validation were generated by side-directed mutagenesis as described previously.17 Our validation confirmed that PSQ is a reliable method to accurately quantify the level of heteroplasmy of the m.13513G>A mutation with an accuracy of±5% and sensitivity (ie detection limit) of 5% (not shown). Although the assay was originally designed to detect the m.13513G>A mutation, the same test also reveals the presence of the m.13514A>G mutation. Figure 1 shows the theoretical pyrograms for these two mutations in case of wild-type mtDNA (Figure 1a) as well as the situation in case of the m.13513G>A mutation (Figure 1b and c) or the m.13514A>G mutation (Figure 1d and e). Differences in relative signal intensities of the first three peaks in the pyrogram compared to the wild-type situation reflects the presence of either one of the mutations. PSQ™96MA Allele Quantification software uses the changes in relative peak height to calculate the level of heteroplasmy. We assume that simultaneous presence of the m.13513G>A and the m.13514A>G mutation in a single patient does not occur. All patient samples were tested using this assay. Confirmation of positive samples was performed by standard automated cycle sequencing of PCR products using an ABI 3730 capillary sequencer and Big Dye Terminator chemistry (Applied Biosystems, Lekkerkerk a/d IJssel, The Netherlands) (Figures 2, 3).
Theoretical prediction of the test results using our PSQ to test for the presence of the m.13513G>A and m.13514A>G mutation in the mitochondrial ND5 gene. The pyrosequence output corresponds to nt. 13512–13520. The sequence to analyze was A(G/A)ACCACAT in case of the m.13513G>A mutation and AG(A/G)CCACAT in case of the m.13514A>G mutation. (a) Expected results in case of 100% wild-type mtDNA molecules, (b) in case of 50% m.13513G>A, (c) 100% m.13513G>A, (d) 50% m.13514A>G and (e) 100% m.13514A>G. It is assumed that simultaneous presence of both mutations in a single patient does not occur.
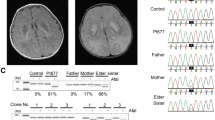
Results of the PSQ in a control blood sample (a) and in (b) blood and (c) muscle mtDNA derived from patient 2. Presence of the m.13513G>A mutation of the mitochondrial DNA is reflected by the increase of the first A peak signal in the pyrogram and simultaneous decrease of the first G and second A signal (peaks within the shaded box). The level of heteroplasmy is calculated by PSQ 96 MA allele quantification software. The levels of heteroplasmy in this patient were 26% in blood and 64% in muscle tissue.
Standard sequence analysis was used to confirm the presence of the m.13513G>A mutation in the ND5 gene in different tissues derived from patients 1 and 2. Samples were initially found to be positive for this mutation by PSQ. The sequence chromatograms shown corresponds to nt. 13512–13520 of the mitochondrial DNA. Levels of heteroplasmy denoted in this figure were assessed by PSQ. The arrowhead marks the site of the mutation.
Results
We screened 113 patients with reduced complex I activity in muscle and 10 patients with deficient complex I activity in fibroblasts for the presence of the m. 13513G>A mutation in the mitochondrial ND5 gene using our PSQ assay. Two patients, both clinically affected with LS, were identified to carry this mutation. Presence of the mutation was confirmed by standard sequence analysis (Figure 3). None of the 123 tested patients harbored the more rare m.13514A>G mutation. Clinical features of the two patients carrying the m.13513G>A mutation are described below and are summarized in Table 1.
The first patient was a Dutch male with LS who died at the age of 23 years. He was born after an uneventful pregnancy and delivery with a normal birth weight and without congenital anomalies. His parents and older sister were healthy. At the age of 18 months developmental delay, strabismus, optical atrophy and spastic tetraparesis were noted. He was diagnosed to have LS at the age of 7 years, with typical lesions on CT-scan and elevated lactate in cerebrospinal fluid. On further examination he appeared to have a Wolff–Parkinson—White syndrome (WPWS)-like cardiac conduction defect and hypercholesterolemia. Later, nystagmus and dysarthria were noted and he was probably mildly mentally retarded (IQ never defined). From the age of 17 years the patient needed peritoneal dialysis because of renal insufficiency, probably caused by dysplasia of the kidneys. At age 22 years he underwent a femoral head resection because of cox arthrosis. The patient died from a septic shock following peritonitis. The first muscle biopsy at the age of 7 years showed no clear abnormalities neither morphologically nor biochemically. The second biopsy 15 years later showed severe reduction in respiratory chain complex I activity with a residual activity of 33% compared to the lowest control value (33 mU/U COX; reference range 101–389) and a mild reduction in complex III activity showing a residual activity of 80% compared to lowest control value (812 mU/U COX; reference range 1020–2530). In fibroblasts, the activities of the respiratory chain enzymes were normal. We detected the m.13513G>A mutation in muscle with a level of heteroplasmy of 63%. Subsequent analysis of a blood DNA sample showed a mutation level of 42%. No muscle from the earlier biopsy was available for mutation analysis.
The second patient was a female diagnosed with LS at the age of 9 years after neuroradiological examination for complaints of headaches, blackouts, diminished visual acuity and ophthalmoplegia. She was the first child of a healthy mother of Norwegian descent and a healthy father of Surinam descent. Pregnancy and delivery were uneventful. Birth weight was normal and no congenital anomalies were noted. Her psychomotor development was slow but she followed mainstream education until the age of 12 years. During childhood there was no progression of neurological disease. Electrocardiography at the age of 9 years revealed a WPWS-like cardiac conduction defect but this was not seen at renewed assessment at the age of 19 years. Renal ultrasound showed a double system of the left kidney with an ureterocele for which she was operated. At the age of 13 years optical atrophy was noted. She is now aged 25 years and the disease has progressed, showing bradyphrenia, ataxia, an extrapyramidal movement disorder, tremor, nystagmus, ptosis, dysarthria and muscle weakness. Biochemical analysis of muscle biopsy material was performed when the patient was 9, 14 and 21 years old, respectively. Only in the last biopsy complex I was clearly deficient displaying a residual activity of 31% compared to the lowest control value (20 mU/U COX; reference range 65–164), while the activities of complex II, III and IV were within the normal range. The reduced complex I activity was in agreement with the strongly reduced pyruvate and malate oxidation rates as well as the clearly reduced ATP production rate of 16 nmol ATP/h.mU citrate synthase (control range 42–81 nmol ATP/h.mU citrate synthase) that were measured in this muscle specimen. Histochemical analysis of the biopsy revealed ragged red fibres. The m.13513G>A transition was found in 64% of mtDNA molecules in muscle. In fibroblasts, collected at the same time as the last muscle biopsy, we measured a residual complex I activity of 85% compared to the lowest control value (93 mU/U COX; control range 110–260 mU/U COX). Subsequent analysis of fibroblast DNA revealed a heteroplasmy level of 25%, which is in agreement with the relatively mildly reduced complex I activity in these cells. In blood, the heteroplasmy level was 26%. No muscle was retained from the earlier biopsies to assess the level of heteroplasmy. The mutation was not detectable in blood from the patient's healthy mother and brother.
Discussion
In the present study we tested a selected panel of 123 patients for the presence of the m. 13513G>A mutation in the mitochondrial ND5 gene which encodes a subunit of respiratory chain complex I. This mutation has previously been identified in patients with LS and in patients with MELAS. All 123 patients exhibited a biochemical complex I deficiency in muscle tissue (n=113) or in fibroblasts (n=10). Of this selected group of patients only two carried the mutation (1.6%). As in one of them the enzyme activity of both complex I and complex III was below the control range, the frequency when considering only isolated complex I deficient cases, would be only one in 68 (1.5%). Previous studies found higher frequencies. Chol et al.6 tested 49 complex I deficient patients of whom 14 clinically had LS. Three of them (6.0%), all with LS, carried the m.13513G>A mutation. Kirby et al.7 detected the mutation in three out of 86 patients (3.5%) with a complex I deficiency, including 30 with LS. All their positive cases also clinically had LS. A Japanese group detected the m.13513G>A transition in six of 84 patients with clinically LS (7.1%), but they found no complex I deficiency in the four young patients in whom this was measured.9 All 12 other mutation positive cases reported in the literature in whom complex I activity was assessed, indeed showed reduced activity.2, 3, 4, 5, 6, 7, 8, 10 Our study shows that the mutation is not as frequent as previously suggested and is most likely to be present in the group of patients with LS together with a complex I deficiency. A complex I deficiency, however, is not always detectable at a young age as is exemplified by the early biopsies at age 7 and 14 years, respectively, in the current two patients. This might be explained by a possible increase in the level of heteroplasmy with time, however, we were unable to test this in our patient since material of the earlier biopsies was no longer available. In addition to our primary cohort consisting of the 123 patients with reduced complex I activity in muscle or fibroblast, which was the main focus of the current study, we also examined an extra panel of 155 independent subjects referred to our diagnostic lab for mtDNA investigation because of clinical suspicion for Leigh (n=58) or MELAS syndrome (n=97) but whose complex I activity status was either normal or unknown. DNA in this patient panel was derived from blood (n=101), muscle (n=46) or fibroblasts (n=8). This cohort also had tested negative for the known mtDNA mutations m.3243A>G, m.8344A>G and m.8993T>G/C and detailed clinical information, other than the suspicion for LS or MELAS, was usually lacking. The m.13513G>A mutation was not found in any of the samples of this latter cohort. This finding corroborates our earlier conclusion that the mutation is not a very frequent cause of Leigh- and MELAS-like syndrome and is most likely to be found in patients with a complex I deficiency.
The clinical presentation of the m.13513G>A transition is highly variable since both MELAS and LS can be caused by this mutation and the age of onset of neurological disease ranges from infancy till the fourth decade. The reason for the clinical variability is unclear. It seems that it cannot be explained by differences in mutation load in muscle since there is overlap between the levels of heteroplasmy found in the early onset severe cases and the milder cases. Mutation loads for this transition, however, seem to differ between different tissues as exemplified in our patients. Therefore, levels in the brain might be different from, and unrelated to, levels in muscle and at least in part explain the different phenotype between patients. Optic atrophy, cataract and deafness are often observed and can be the first sign of the disease; long before CNS involvement becomes apparent. We confirm that there is a relationship between severe involvement of the optic nerve and mutations causing amino-acid changes at position D393 in the ND5 protein.4, 10, 11 Sudo et al.9 postulated that a WPWS-like cardiac conduction defect is typical of the m.13513G>A mutation since this was described in three of their six patients, whereas two had another cardiac conduction abnormality. Our two cases corroborate this hypothesis. WPWS, however, is occasionally seen in other mitochondrial encephalomyopathies and in patients with Leber hereditary optic neuropathy.19, 20 It is remarkable that both our cases displayed renal abnormalities, however, it remains unclear whether this should be regarded as part of the m.13513G>A phenotype since renal abnormalities have not been described in connection with this mutation before. The same holds for the hypercholesterolemia of the first patient, particularly since this feature has recently been associated with a mtDNA mutation in a large multigeneration family.21 Despite the great variability in clinical presentation, optical atrophy and WPWS seem quite specific and frequent features in patients with the m.13513G>A transition.
Thus far, only four patients have been reported with the m.13514A>G transition, the second mutation we tested in this study.4, 22 The fact that we found no cases with this mutation confirms that it indeed appears a rare cause of complex I deficiency, LS or MELAS syndrome.
We have shown that PSQ is a rapid and efficient method for detection of the m.13513G>A and m.13514A>G transitions, and for determination of the level of heteroplasmy. Classical PCR-restriction digestion (RFLP) approaches are effective in assessing presence of the mutation, however, they are time consuming and less reliable in determining the level of heteroplasmy.
In summary, we conclude that the m.13513G>A mutation is an infrequent cause of LS and a rare cause of MELAS. Screening for this mutation as part of the routine diagnostic workup of all referrals received for mtDNA analysis is not indicated. We do recommend screening for the m.13513G>A transition in patients with Leigh (-like) syndrome with complex I deficiency or optic atrophy and/or WPWS. We confirm that pyrosequencing is an effective method for quick and reliable screening of this mutation.
References
Smeitink J, van den Heuvel L, DiMauro S : The genetics and pathology of oxidative phosphorylation. Nat Rev Genet 2001; 2: 342–352.
Santorelli FM, Tanji K, Kulikova R, Shanske S, Vilarinho L, Hays AP, DiMauro S : Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochem Biophys Res Commun 1997; 238: 326–328.
Penisson-Besnier I, Reynier P, Asfar P et al: Recurrent brain hematomas in MELAS associated with an ND5 gene mitochondrial mutation. Neurology 2000; 55: 317–318.
Corona P, Antozzi C, Carrara F et al: A novel mtDNA mutation in the ND5 subunit of complex I in two MELAS patients. Ann Neurol 2001; 49: 106–110.
Pulkes T, Eunson L, Patterson V et al: The mitochondrial DNA G13513A transition in ND5 is associated with a LHON/MELAS overlap syndrome and may be a frequent cause of MELAS. Ann Neurol 1999; 46: 916–919.
Chol M, Lebon S, Benit P et al: The mitochondrial DNA G13513A MELAS mutation in the NADH dehydrogenase 5 gene is a frequent cause of Leigh-like syndrome with isolated complex I deficiency. J Med Genet 2003; 40: 188–191.
Kirby DM, Boneh A, Chow CW et al: Low mutant load of mitochondrial DNA G13513A mutation can cause Leigh's disease. Ann Neurol 2003; 54: 473–478.
Petruzzella V, Di Giacinto G, Scacco S et al: Atypical Leigh syndrome associated with the D393N mutation in the mitochondrial ND5 subunit. Neurology 2003; 61: 1017–1018.
Sudo A, Honzawa S, Nonaka I, Goto Y : Leigh syndrome caused by mitochondrial DNA G13513A mutation: frequency and clinical features in Japan. J Hum Genet 2004; 49: 92–96.
Bugiani M, Invernizzi F, Alberio S et al: Clinical and molecular findings in children with complex I deficiency. Biochim Biophys Acta 2004; 1659 (2–3): 136–147.
Liolitsa D, Rahman S, Benton S, Carr LJ, Hanna MG : Is the mitochondrial complex I ND5 gene a hot-spot for MELAS causing mutations? Ann Neurol 2003; 53: 128–132.
Budde SM, van den Heuvel LP, Smeitink JA : The human complex I NDUFS4 subunit: from gene structure to function and pathology. Mitochondrion 2002; 2 (1–2): 109–115.
Bentlage HA, Wendel U, Schagger H, ter Laak HJ, Janssen AJ, Trijbels JM : Lethal infantile mitochondrial disease with isolated complex I deficiency in fibroblasts but with combined complex I and IV deficiencies in muscle. Neurology 1996; 47: 243–248.
Smeitink J, Sengers R, Trijbels F, van den Heuvel L : Human NADH: ubiquinone oxidoreductase. J Bioenerg Biomembr 2001; 33: 259–266.
Ronaghi M : Pyrosequencing sheds light on DNA sequencing. Genome Res 2001; 11: 3–11.
White HE, Durston VJ, Seller A, Fratter C, Harvey JF, Cross NC : Accurate detection and quantitation of heteroplasmic mitochondrial point mutations by pyrosequencing. Genet Test 2005; 9: 190–199.
Biggin A, Henke R, Bennetts B, Thorburn DR, Christodoulou J : mutation screening of the mitochondrial genome using denaturing high-performance liquid chromatography. Mol Genet Metab 2005; 84 (1): 61–74.
Lowik MM, Hol FA, Steenbergen EJ, Wetzels JF, van den Heuvel LP : Mitochondrial tRNALeu(UUR) mutation in a patient with steroid-resistant nephrotic syndrome and focal segmental glomerulosclerosis. Nephrol Dial Transplant 2005; 20 (2): 336–341.
Mashima Y, Kigasawa K, Hasegawa H, Tani M, Oguchi Y : High incidence of pre-excitation syndrome in Japanese families with Leber's hereditary optic neuropathy. Clin Genet 1996; 50: 535–537.
Nikoskelainen EK, Savontaus ML, Huoponen K, Antila K, Hartiala J : Pre-excitation syndrome in Leber's hereditary optic neuropathy. Lancet 1994; 344: 857–858.
Wilson FH, Hariri A, Farhi A et al: A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science 2004; 306: 1190–1194.
Lebon S, Chol M, Benit P et al: Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J Med Genet 2003; 40: 896–899.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ruiter, E., Siers, M., van den Elzen, C. et al. The mitochondrial 13513G>A mutation is most frequent in Leigh syndrome combined with reduced complex I activity, optic atrophy and/or Wolff–Parkinson–White. Eur J Hum Genet 15, 155–161 (2007). https://doi.org/10.1038/sj.ejhg.5201735
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201735
Keywords
This article is cited by
-
Induced pluripotent stem cells derived from patients carrying mitochondrial mutations exhibit altered bioenergetics and aberrant differentiation potential
Stem Cell Research & Therapy (2023)
-
Enhanced mitochondrial DNA editing in mice using nuclear-exported TALE-linked deaminases and nucleases
Genome Biology (2022)
-
Molecular basis of Leigh syndrome: a current look
Orphanet Journal of Rare Diseases (2020)
-
Multisystem mitochondrial diseases due to mutations in mtDNA-encoded subunits of complex I
BMC Pediatrics (2020)
-
Mitochondrial DNA 11777C>A Mutation Associated Leigh Syndrome: Case Report with a Review of the Previously Described Pedigrees
NeuroMolecular Medicine (2010)