Abstract
The circadian clock percolates through every aspect of behaviour and physiology, and has wide implications for human and animal health. The molecular basis of the Drosophila circadian clock provides a model system that has remarkable similarities to that of mammals. The various cardinal clock molecules in the fly are outlined, and compared to those of their actual and ‘functional’ homologues in the mammal. We also focus on the evolutionary tinkering of these clock genes and compare and contrast the neuronal basis for behavioural rhythms between the two phyla.
Similar content being viewed by others
Introduction: clocks and disease
The number of reviews written on biological rhythms in the past 15 years has been enormous, particularly those on the molecular aspects. So, why are we writing another one on Drosophila, and why for a readership of human/medical geneticists who must care little or nothing for such a subject or such an organism? After all, 24 h circadian rhythms, on which most ‘chronobiologists’ focus, have little or nothing to do with human disease, and Drosophila are of no interest to such an audience anyway given their vast phylogenetic distance from mammals. The answer to the question is two-fold. One is that the editors used their arsenal of weapons, flattery and charm, to convince the reluctant senior (reflecting age) author (CPK) to do this piece. The second reason is more interesting, in that we believe that medical geneticists should know about circadian rhythms and their links to disease, and should know that Drosophila has provided the model organism on which almost all the major clock genes were first identified. Within this latter sentence lies the implicit understanding that the genes that control the fly circadian clock are the same ones that determine the corresponding human 24 h cycle.
Is there a relationship between circadian clocks and disease? In Western societies, about 20% of the population, perhaps more, work in shifts. There are various types of shift-work programmes, but all have the effect of desynchronising the workers internal clock to the outside world. The effects of chronic shift-work have been documented in numerous studies, and in terms of general health and life expectancy, there is nothing to commend it. Shift workers suffer elevated levels of almost everything negative that has been investigated, including sleep, gastrointestinal and cardiac problem, even cancer.1 While some of these investigations have yet to be duplicated, there is clearly an anxiety about the effects of shift-work on health.
However, there are other reasons for human/medical geneticists to be interested in fly clocks. Over the past few years, a number of papers have appeared revealing that flies appeared to sleep in a manner that was similar to mammals.2, 3, 4 This immediately opened up the traditional area of sleep research to mutational analysis, and at least two clock-related transcription factors, CREB and CYC, have been shown to play important roles in sleep homeostasis.3, 5 In addition, another area in which fly clock genes, and a subset of the neurons in which clock genes are expressed, is important, is the behavioural response to cocaine administration.6, 7 Thus, the molecular genetic analysis of the fly's circadian system may play a useful role in understanding one clearly clock-related phenotype, and another in which the relevance of clock phenomena is not quite so immediately obvious. Nevertheless, there is one particular human syndrome, where circadian clocks have been directly linked.
Familial advanced sleep phase syndrome (FASPS)
There are individuals within families that apparently carry a dominant gene that causes them to wake up very early, and go to bed early8. This is reminiscent of the classic pers mutant in Drosophila, where flies have a 19 h rhythm9 and their locomotor activity occurs a few hours earlier than in the wild-type fly, when they are placed in a 12 h light: 12 h dark (LD12:12) schedule. The pers mutation is a serine-to-asparagine change in that part of the PER protein that interacts with a kinase called Doubletime10, 11 (aka casein kinase 1ɛ, CK1ɛ, in mammals). When the FASPS gene was identified in an extensive single-family pedigree, imagine the excitement when it was discovered that the mutation mapped to the human Per2 gene.8 Per2 is one of four Per genes in mammals, the first three being transcribed and translated, the fourth being a pseudogene.12, 13 The human Per2 locus is the one that is closest to the fly's single per, suggesting it is the ancestral version from which the others duplicated.12 Imagine the further excitement when the lesion in hPer2 was discovered to be within a serine codon, which was replaced by glycine in the FASPS individuals, in the CK1ɛ-interacting region. In other words, the first identified clock mutation in human and fly was in the same gene, in a target site for CK1! However, more recently, a second set of families with FASPS has been discovered. This time, the lesion is not in hPer2, but in CK1δ,14 which is also the homologue of fly DBT, as flies only have one version of this kinase gene. Both per and doubletime (CK1ɛ) were first identified via mutant screens in flies,9, 15 revealing how Drosophila can lead the way in human gene discovery, and giving ‘fly guys’ like us, a further means to justify our grant proposals.
Clock genes in fly (and mouse): a very brief historical perspective
Flies do not always lead mammals however. Joe Takahashi's group identified the Clock mutant in the mouse in a mutagenesis in 1994,16 and cloned it in 1997.17, 18 A little later, the gene, Jerk, was identified in Drosophila, again by forward genetics, and the DNA sequence revealed it to be the homologue of mammalian Clk.19 The fly mutant has since been renamed ClkJrk. However, apart from this single case, most of the core clock genes, which are shared between flies and mammals, were identified initially in the fly.
The first, and most famous study was that of Konopka and Benzer,9 in which a chemical mutagenesis of the fly X chromosome identified pers, and two other alleles perl (29 h rhythm) and per0 (arrhythmic). The initial character for the screen was pupal–adult eclosion of fly populations, taking advantage of the fact that wild-type flies eclose around dawn. This reflects their adaptation to their ancestral homeland of sub-Saharan Africa, where dawn has the most humid conditions for pumping haemolymph into the wings. Eclosing in the dessicating conditions of the afternoon could be disastrous (ie these late eclosing fruit-flies would be ‘fruit-walks’). Indeed, Drosophila means ‘dew-lover’ in Greek, so whoever named them was a genius. However, this population circadian assay, which gives bursts of eclosion every 24 h, even in constant conditions, is of little use where the investigator is interested in the genotype of individual flies. For this, Konopka and Benzer moved to locomotor activity assays, as individual flies show circadian patterns of sleep and wakefulness, just as in humans. In retrospect, it can be argued that Konopka and Benzer identified the first dedicated ‘behavioural’ gene in any higher organism, as without the per gene, flies seemed not to suffer serious viability problems, they are simply arrhythmic. However per does have pleiotropy associated with it which includes other effects on temporal programmes that are not circadian,20, 21, 22 as well as phenotypes that do not seem to have anything to do with the dimension of time.23
DNA around the per locus was cloned in mid-1984, but putative per sequences were functionally assayed by transformation in December of that year.24, 25 The first behavioural gene identified by forward genetics had been isolated………a big deal in those days. It took 10 more years for the next clock gene to come on board, courtesy of Michael Young's group, and that was timeless, an autosomal locus.26 In 1995, tim was cloned,27 and it was simultaneously discovered that TIM protein could physically associate with PER,28 via the PER PAS domain, a sequence bearing two imperfect 51-amino-acid repeats separated by a long stretch of >150 residues.29 In fact, PER is one of the three founder members of the PAS domain family, the other two being transcription factors, ARNT (Aryl hydrocarbon nuclear translocator) and SIM (single-minded). Since the discovery of PAS, a huge family of PAS proteins have been discovered, from bacteria to eukaryotes,30, 31 a subset of which are responsive to light, oxygen and voltage giving rise to PAS LOV proteins.32 As the ancestral clock proteins must have responded to light cycles, could PAS be the protein module that links extant clock proteins with the PASt? Maybe, but Cryptochrome, the dedicated circadian blue-light photoreceptor in flies (see below), which is related by ancestry to the photolyases, ancient DNA-binding proteins that repair UV-induced damage, may also have played a starring role in the evolution of the clock mechanism below the oceans, where blue light can penetrate.33
Cycling gene products and negative feedback
The products of the per and tim genes, RNA and proteins, cycle in abundance in the fly's head in constant conditions with a few hours lag between the RNA peak and the protein's.34, 35, 36, 37 As RNA goes up, protein comes down and vice versa, suggesting a negative feedback loop. Consistent with this, both PER and TIM proteins are seen at night, but in a limited number of neurons in the brain. When the proteins are at their peak of abundance late at night, the two proteins move into the nuclei of the various subsets of clock neurons,37, 38, 39, 40 where PER (but not TIM) is believed to act as the major negative regulator.41 In all, there are about 150 of these master clock neurons, and they provide a much simpler model system than the mammalian suprachiasmatic nuclei (SCN), which houses the corresponding neuronal clock and contains about 20 000 clock cells.42
The PER and TIM proteins, therefore, wax and wane in a circadian cycle, and once inside the nucleus, they interact with two bHLH PAS proteins, CLOCK, which we mentioned before, and CYCLE, both defined by mutagenesis.19, 43 CYCLE is the homologue of BMAL1 in mammals, and both play similar roles in the clock. The CLK-CYC dimer activates transcription by binding to CACGTG E-boxes in the per and tim promoter regions.44 Thus, the CLK-CYC dimer forms the positive limb of the feedback loop and when PER enters the nucleus, it interacts with the dimer (incidentally, not via PER PAS as had been commonly assumed, but via a C-terminal region), and represses transcription.45 PER eventually degrades via the phosphorylation of DBT15, 46 (although phosphorylation by CK247, 48 is also important for shifting PER from the cytoplasm to the nucleus), and transcription is reactivated. The delay between per mRNA and protein accumulation is also due to DBT.49 However, DBT protein does not cycle, but the dephosphorylation of PER does, and is mediated by cycling expression of the relevant phosphatase, PP2A.50 As PER is translated in the cytoplasm in the early night, it is phosphorylated by DBT as a prelude to degradation, in which the product of slimb (supernumery limbs), a ubiquitin ligase, becomes critical.51, 52 It is only with the build-up of TIM, itself phosphorylated by the product of shaggy, aka GSK3,53 that this premature degradation of PER ceases, and PER stabilises, and moves into the nucleus late at night with the help of TIM.54 PER and TIM mutually stabilise and cooperate to ensure each other's movement and retention within the different cellular compartments.55 However, they do not appear to be obligatory partners in vivo for nuclear localisation,56 as initially believed based on ex in vivo experiments with cell lines.57
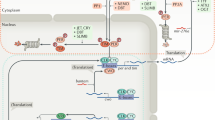
This feedback loop of two positive, two negative and a number of modulatory components (DBT, SGG, SLIMB) is coupled to a second feedback loop via CLK. CLK, unlike CYC, oscillates in abundance.58 CLK/CYC activates the transcription of two genes vrille, and Pdp1ɛ (Par domain protein 1ɛ) which encode transcription factors, with VRI (negative) and PDP1ɛ (positive) acting in opposite ways (see Figure 1) to regulate the transcription of Clk.59 Coupling two loops in this way enhances the stability of both cycles, and it remains to be seen whether there are further feedback loops entrenched within this basic system.
Two interlocked feedback loops in the Drosophila clock. The per and tim genes are negative regulators of their own transcription via the CLK/CYC transcription factors. The Clk gene is interlocked with the per/tim loop via the positive and negative actions of the transcription factors PDP1ɛ and VRI, respectively.
How the clock responds to light
The circadian feedback loops must be responsive to the external environment, particularly light and heat. One way that this is achieved for light is through the canonical visual pathway mediated by the rhodopsins. The other way is through the cryptochrome (cry) gene that encodes a blue-light-sensitive molecule mentioned earlier. CRY is activated by light, and binds to both TIM and PER60, 61 in transient interactions that are difficult to capture in vivo.62 A consequence of the activation of CRY is that it precedes light-induced TIM ubiquitination and degradation.63 A nearly null mutant, cryb, does not show any alteration in free-running circadian rhythms under DD, but does have drastically reduced circadian phase responses to short light pulses,64 revealing its function for light signalling to the clock.65, 66 However, with long light pulses, as in LD12:12 cycles, cryb flies entrain relatively normally, reflecting the underlying function of the rhodopsins.64 The light response of TIM provides the basis for how a clock entrains to different light regimens.67, 68 Consider that early at night, when tim mRNA levels are high, but TIM levels low, the reduction of TIM levels by a light pulse (via CRY signalling) takes the clock backwards a few hours to a phase that had less TIM.37, 69, 70 However, the levels of tim mRNA restore TIM within a few hours, effectively generating a time delay for the clock. The same light stimulus late at night reduces the high levels of TIM to the level that they will be in a few hours time, that is, low TIM, but as there is little tim mRNA, the move to a future time (with less TIM) is permanent, giving a phase advance.37, 69, 70 This compelling (but perhaps superficially understood mechanism), explains beautifully the circadian light phase response, where light pulses early at night give delays and those late at night give advances, and is a universal feature of circadian clocks. The delay between tim mRNA and protein provides the pivotal feature for explaining this mechanism.
Central and peripheral clocks in flies
So far, we have dealt only with clock gene expression in the brain, which controls the rhythmic behavioural output that we observe. However, autonomous light-sensitive clocks are almost everywhere in the fly, as seen by fusing luciferase sequences to the per promoter region. In these beautiful experiments, the fruitfly becomes a cycling firefly, with rhythms of luciferase observed in isolated legs, eyes, proboscis, wings, antennae, etc.71 Thus, compared to mammals, there are autonomous, independent light-regulated clocks in the fly. In the mouse, for example, cycling in peripheral tissues depends on the master clock, the SCN, so that in the absence of SCN signalling, peripheral clocks become desynchronised and rapidly dampen.72, 73 However, this traditional idea of the SCN being the master oscillator may need to be amended because recent reporter-clock protein fusions reveal that peripheral tissues can maintain robust rhythms in the absence of the SCN, although synchrony between tissues and between animals may be lost.74
In the fly, in at least two of these peripheral clocks, the antennae and the eyes, the cryb mutation appears to stop the clock,64, 75 whereas it has little effect on free-running circadian locomotor behaviour, which is generated from the clock neurons mentioned above. Thus, CRY is a flexible molecule that plays a light-sensing role that can modulate the central ‘behavioural’ clock, but does not disrupt it when mutated, whereas in the periphery, it appears to encode a more fundamental clock feature. Interestingly, in mammals, CRY is the major negative regulator of the circadian clock, playing very much the PER/TIM role, so the central clock of mammals is functionally similar to the peripheral clock of flies. This evolutionary tinkering of the role of CRY between mammals and flies is also seen in the fact that CLK cycles in flies,58 whereas CYC does not, but this role is reversed for mammalian CLK and BMAL1.76 In addition, the coupling of the PER/TIM and CLK feedback loops mentioned earlier, via VRI and PDP1ɛ, is also observed in mammals, in which BMAL1 cycling is determined by the balance between Rev-erbα and RORa.77, 78
Further evolutionary tinkering: gene duplication and divergence
From the evolutionary point of view, one of the striking differences between mammalian and fly clocks is that vertebrates seem to employ multiple paralogous clock genes, while the Drosophila clock usually does not involve duplicated genes.79, 80 For example, the mammalian clock has at least two cry genes or paralogues (the zebra fish has six), three functional paralogues of per, two of Clock and two of Bmal1.80 In contrast, in Drosophila, all these genes exist in one copy only. This is apparently a general feature resulting from the genome expansion (duplication) during vertebrate evolution. Once gene duplication occurs, the selective constraints on the new copy are reduced, allowing the new copy (or both) to evolve and to acquire a new function resulting in functional divergence.81 For example, the three functional mouse Per paralogues appear to play different roles.82, 83, 84 A recent study suggested that mPER1 is important for prolonged seasonal light adaptation,85 while mPER2 was previously shown to be important in generating the daily activity cycles.84
In Drosophila, only one per gene exists, but this does not apply to all insects because in the silk moth, Antheraea pernyi, two paralogues are present perZ and perW on the different sex chromosomes.86 The rhythmicity of the males, which are the homogametic sex (chromosomes ZZ), suggests that perW is not crucial for circadian function. Many additional degenerate per sequences exist on the female W chromosome including one that generates an intriguing antisense transcript.86
In one case, however, a Drosophila circadian clock gene, timeless, does have a paralogue called timeout (aka tim2). Although it is not yet clear whether tim2 has a circadian function in the fly, this sequence is more closely related to the single mammalian tim sequence, than is fly tim.87, 88 Thus, tim2 must be the ancestral sequence, which in diptera and lepidoptera, at least, has relatively recently duplicated to generate the clock-relevant tim. A role for mTim in the mammalian clock has been proposed from the work of Barnes et al89 and Ünsal-Kaçmaz et al,90 the latter suggesting that the role of mammalian TIM is to couple the circadian clock to the cell cycle. This relates well with the observation that tim has sequence similarity to cell-cycle-related proteins, for example, the yeast Sw1 (Switching deficient)91 and Tof1 (Topoisomerase 1-interacting factor),92 which are involved in DNA damage activation checkpoints. In addition, TIM1 from Caenorhabditis elegans is important for chromosome cohesion.93 However, any role of mTIM in the mammalian clock would appear to be very different from the light-sensitive negative regulator function of fly TIM.
TIM proteins are intriguing, in that they are pioneer proteins and do not look much like anything else in the databases. By using various conformational algorithms, a number of papers have suggested that TIM may contain ARM domains,93, 94, 95, 96 all α-helical repeat motifs found in proteins such as fly ARMADILLO (aka β-catenin in mammals). While the bases for these types of bioinformatic analyses have been hotly disputed,97 ARMADILLO/β-catenin is the focal point for the wingless (or Wnt) signalling pathway, which converges on the ARM/β-catenin transcription factor to regulate its stability and subsequent nuclear accumulation/retention. If TIM is an ARM-like protein, could this explain why so many of the wingless pathway members have been recruited into the clock mechanism, DBT, SGG, SLIMB, CK1ɛ, CK2, PP2A (see above)?
Role of splicing in the fly clock
Splicing may provide another route to achieve fine-tuning of a system, and may serve a similar function as gene duplication, with the different splice isoforms undergoing functional divergence. Kopelman et al98 found that gene duplication and splicing are inversely correlated in the human and mouse genomes, with large gene families showing little alternative splicing, whereas single-copy genes with no paralogues had high levels of alternative splicing. In Drosophila, the per 3′UTR undergoes alternative splicing, which is regulated by temperature and light, and determines seasonally adaptive changes in the timing of the fly's locomotor activity.99, 100, 101 Intriguingly, it is the act of alternative splicing per se that is important, not the informational content of the two per 3′ splice forms that is important. At colder temperatures, splicing stimulates the earlier upswing of per mRNA, leading to earlier behavioural activity.101 The implications of this is that at hotter temperatures, the fly's main burst of activity moves to later, cooler parts of the day, thereby avoiding the ‘fruit-walk’ dessication scenario outlined earlier. Analysis of the fly genome (www.flybase.org) suggests that other clock genes have multiple splicing isoforms, for example, vri has two isoforms, tim has five and Clk has six. We feel a PhD project or two beckoning us here!
The ‘neuro’ revolution in chronobiology: anatomy of the clock
In the last few years, rather than grinding up the fly head and running Western or Northern blots, workers have started to dissect the contributions of the different clock neurons to circadian behavioural output. Figure 2 shows these cells, which constitute the central clock. There are six main clusters that stain strongly with PER and TIM antibodies. These are divided in three groups of lateral neurons (LNs) and three groups of dorsal neurons (DNs).39, 40, 102, 103, 104, 105 The LNs are further subdivided into dorsal (LNd – one cluster of ∼6 cells) and ventral, the latter further subdivided into four large (l-LNv) and four small (s-LNv) neurons. These LNv neurons are usually missing in disconnected mutants, which are behaviourally arrhythmic.40, 104 Furthermore, these LNv cells also produce the neuropeptide PDF,102, 105 shown to be essential not for rhythmicity per se, but as a synchronising signal among different classes of clock neurons.106, 107, 108 The DNs, which are located more posteriorly, are subdivided into DN1 (∼15), DN2 (2) and DN3 (∼40 cells in a very dorsolateral position).109, 110, 111
Central clock cells. Confocal projection of optical slices (z-axis) from a Drosophila melanogaster brain sampled at the end of the night (ZT 0) and stained with anti-Per antibody. Anti-PER antibody135 was used at a dilution of 1:15 000. Goat anti-rabbit IgG-Cy3 (Jackson, 1:200) was used for detection. The size bar is represents 100 μm. The major clock clusters are all visible. l-LNvs=large lateral neurons ventral; s-LNvs=small lateral neurons ventral; LNd=dorsal lateral neurons; DN1=dorsal neurons 1; DN2=dorsal neurons 2; DN3=dorsal neurons 3; and Me=medulla.
The wiring of the circadian network
Evidence that clock neurons are not equivalent and do not act in isolation lies initially in the architecture of their neuronal projections.109, 110, 111 With the exception of the l-LNvs, most of the clock neurons project to a wide area of the dorsal brain where we find most of the fly neurosecretory cells, as well as integration and associative regions for motor behaviour.110 However, the main circadian centre is probably the accessory medulla, a mass of neuronal processes derived from the larval optic lobe.112, 113 The s-LNvs lay at the edge of the accessory medulla from where they extend their dendritic arborisation.112 Further input to the accessory medulla comes from the Hofbauer–Buchner eyelet, an extraretinal photoreceptor derived from the visual (Bolwig's organ) system of the larval,112 and, via the l-LNvs, from the eyes.110 The s-LNvs project to an area of the brain in very close proximity to the DN1s and DN2s,110, 111 possibly communicating with these cells via PDF and/or some other neurotransmitter. Furthermore, in Pdf0 flies maintained under DD, PER cycling becomes accelerated and dampened in the LNds, suggesting that the s-LNvs use PDF to entrain the LNds in LD conditions.108
Functional differences in circadian cells
The wiring of the clock cells suggests that the s-LNvs are probably the most important cluster in the circadian network. It follows that the physical or functional ablation of the s-LNvs should result in dramatic behavioural effects at least in terms of locomotor activity. Mutant Pdf0 flies, although rhythmic under LD, lack the morning locomotor activity peak, whereas the evening peak is anticipated by about 1 h.106 In DD, the mutant flies remain rhythmic for about 2–3 days, suggesting that this is the synchronisation among clock cells, rather than core clock properties that are affected.106 The physical ablation of the LNvs by overexpression of apoptotic genes has basically the same effect, suggesting that it is via PDF alone that these cells are necessary for sustained rhythmicity in DD and for the morning peak in LD.106 PDF is packed in dense core vesicles localised in varicosities that lack synaptic specialisation, suggesting a nonsynaptic paracrine release of the peptide.114 In agreement with these data, suppression of synaptobrevin-mediated synaptic transmission does not result in a rhythmic phenotype.115
At this point, one should mention that the mammalian circadian system also has a neuropeptidergic axis that shows similar features to the fly's PDF. Vasoactive intestinal peptide (VIP), and pituitary adenylate cyclase-activating peptide (PACAP), may be important in the light entrainment pathway of the mammalian clock.116, 117, 118 VIP is synthesised in retinally innervated, ventrolateral neurons of the SCN,119 whereas PACAP is made in the melanopsin-containing photoreceptor cells of the retina that also innervate the SCN.120 Both peptides use the G-protein-coupled VPAC2 receptor, and receptor knockout mutant mice show various types of circadian behavioural and molecular dysfunction.121 Subsequent analyses reveal that neurons in the SCN may retain cycling clock gene expression, but the absence of the VPAC2 receptor means that they are desynchronised with respect to each other.122 Thus, VIP may, as with PDF in the LNs of the fly, represent a paracrine signal that maintains temporal order between SCN neurons.
Recently, using a variety of techniques that allow the cell-specific manipulation of clock genes, or the ablation of subsets of clock neurons, two groups have shown that the LNvs (actually the s-LNvs) are necessary and sufficient for sustained rhythmicity in DD.123, 124 However, only the morning, not the evening activity component is rescued, both under LD and DD by the s-LNvs. Extending the expression of per also to the LNds, additionally rescues the evening peak,124 and suggests that the s-LNv and the LNds, respectively, govern the morning and evening bouts of locomotion.123, 124 This is an important result as it shows that different parts of the neuronal circadian network control different aspects of the circadian programme as in mammals,125, 126 and suggests an anatomical substrate to the morning (M) and evening (E) oscillator hypothesis first proposed nearly 30 years ago.127, 128
Future directions
We have already discussed PDF as an ‘output’ molecule of the clock that serves to synchronise the various clock neurons. However, there are hundreds of clock gene outputs that serve to generate the behavioural and physiological rhythms that can be observed in flies, and these have been detected using microarray approaches.129, 130, 131, 132, 133 Of course, we do not know if mRNA cycling is accompanied by protein cycling, and future studies will inevitably add the proteomic dimension to the transcriptomic. It may be that many genes cycle at the protein level via post-transcriptional mechanisms without having a cycling mRNA. This remains to be seen. However, the systems approach, as such ‘omic’ programmes are called, have revealed that almost every physiological function has a cycling component in flies and in mammals.72 Our own feeling is that the molecular and physiological dissection of identified neurons, and the determination of their contributions to rhythmic behaviour and entrainment via light, heat and even social stimuli,134 will probably provide the most important growth area for fly clock molecular genetics in the next 5 years.
References
Hastings MH, Reddy AB, Maywood ES : A clockwork web: circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci 2003; 4: 649–661.
Shaw PJ, Cirelli C, Greenspan RJ, Tononi G : Correlates of sleep and waking in Drosophila melanogaster. Science 2000; 287: 1834–1837.
Shaw PJ, Tononi G, Greenspan RJ, Robinson DF : Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 2002; 417: 287–291.
Hendricks JC, Finn SM, Panckeri KA et al: Rest in Drosophila is a sleep-like state. Neuron 2000; 25: 129–138.
Hendricks JC, Williams JA, Panckeri K et al: A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci 2001; 4: 1108–1115.
Tsai LT, Bainton RJ, Blau J, Heberlein U : Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol 2004; 2: e408.
Andretic R, Chaney S, Hirsh J : Requirement of circadian genes for cocaine sensitization in Drosophila. Science 1999; 285: 1066–1068.
Toh KL, Jones CR, He Y et al: An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 2001; 291: 1040–1043.
Konopka RJ, Benzer S : Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 1971; 68: 2112–2116.
Yu Q, Jacquier AC, Citri Y, Hamblen M, Hall JC, Rosbash M : Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci USA 1987; 84: 784–788.
Rothenfluh A, Abodeely M, Young MW : Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr Biol 2000; 10: 1399–1402.
Clayton JD, Kyriacou CP, Reppert SM : Keeping time with the human genome. Nature 2001; 409: 829–831.
Gotter AL, Reppert SM : Analysis of human Per4. Brain Res Mol Brain Res 2001; 92: 19–26.
Xu Y, Padiath QS, Shapiro RE et al: Functional consequences of a CKI delta mutation causing familial advanced sleep phase syndrome. Nature 2005; 434: 640–644.
Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW : double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998; 94: 83–95.
Vitaterna MH, King DP, Chang AM et al: Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 1994; 264: 719–725.
Antoch MP, Song EJ, Chang AM et al: Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell 1997; 89: 655–667.
King DP, Zhao Y, Sangoram AM et al: Positional cloning of the mouse circadian clock gene. Cell 1997; 89: 641–653.
Allada R, White NE, So WV, Hall JC, Rosbash M : A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 1998; 93: 791–804.
Kyriacou CP, Hall JC : Circadian rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the male's courtship song. Proc Natl Acad Sci USA 1980; 77: 6729–6733.
Kyriacou CP, Oldroyd M, Wood J, Sharp M, Hill M : Clock mutations alter developmental timing in Drosophila. Heredity 1990; 64 (Part 3): 395–401.
Beaver LM, Giebultowicz JM : Regulation of copulation duration by period and timeless in Drosophila melanogaster. Curr Biol 2004; 14: 1492–1497.
Beaver LM, Gvakharia BO, Vollintine TS, Hege DM, Stanewsky R, Giebultowicz JM : Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc Natl Acad Sci USA 2002; 99: 2134–2139.
Bargiello TA, Jackson FR, Young MW : Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature 1984; 312: 752–754.
Zehring WA, Wheeler DA, Reddy P et al: P-element transformation with period locus DNA restores rhythmicity to mutant, arrhythmic Drosophila melanogaster. Cell 1984; 39: 369–376.
Sehgal A, Price JL, Man B, Young MW : Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 1994; 263: 1603–1606.
Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A : Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 1995; 270: 805–808.
Gekakis N, Saez L, Delahaye-Brown AM et al: Isolation of timeless by PER protein interaction: defective interaction between timeless protein and long-period mutant PERL. Science 1995; 270: 811–815.
Huang ZJ, Edery I, Rosbash M : PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature 1993; 364: 259–262.
Pellequer JL, Brudler R, Getzoff ED : Biological sensors: more than one way to sense oxygen. Curr Biol 1999; 9: R416–R418.
Pellequer JL, Wager-Smith KA, Kay SA, Getzoff ED : Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA 1998; 95: 5884–5890.
Crosson S, Rajagopal S, Moffat K : The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry 2003; 42: 2–10.
Gehring W, Rosbash M : The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol 2003; 57 (Suppl 1): S286–S289.
Hardin PE, Hall JC, Rosbash M : Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990; 343: 536–540.
Edery I, Zwiebel LJ, Dembinska ME, Rosbash M : Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA 1994; 91: 2260–2264.
Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers MP, Young MW : Rhythmic expression of timeless: a basis for promoting circadian cycles in period gene autoregulation. Science 1995; 270: 808–810.
Myers MP, Wager-Smith K, Rothenfluh-Hilfiker A, Young MW : Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 1996; 271: 1736–1740.
Curtin KD, Huang ZJ, Rosbash M : Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 1995; 14: 365–372.
Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC : Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron 1988; 1: 141–150.
Zerr DM, Hall JC, Rosbash M, Siwicki KK : Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci 1990; 10: 2749–2762.
Rothenfluh A, Young MW, Saez L : A TIMELESS-independent function for PERIOD proteins in the Drosophila clock. Neuron 2000; 26: 505–514.
Hastings M, Maywood ES : Circadian clocks in the mammalian brain. Bioessays 2000; 22: 23–31.
Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC : CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell 1998; 93: 805–814.
Lee C, Bae K, Edery I : PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol Cell Biol 1999; 19: 5316–5325.
Chang DC, Reppert SM : A novel C-terminal domain of Drosophila PERIOD inhibits dCLOCK : CYCLE-mediated transcription. Current Biology 2003; 13: 758–762.
Kloss B, Price JL, Saez L et al: The Drosophila clock gene double-time encodes a protein closely related to human casein kinase I epsilon. Cell 1998; 94: 97–107.
Akten B, Jauch E, Genova GK et al: A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 2003; 6: 251–257.
Lin JM, Kilman VL, Keegan K et al: A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 2002; 420: 816–820.
Bao S, Rihel J, Bjes E, Fan JY, Price JL : The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci 2001; 21: 7117–7126.
Sathyanarayanan S, Zheng X, Xiao R, Sehgal A : Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 2004; 116: 603–615.
Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F : The F-box protein slimb controls the levels of clock proteins period and timeless. Nature 2002; 420: 178–182.
Ko HW, Jiang J, Edery I : Role for Slimb in the degradation of Drosophila period protein phosphorylated by Doubletime. Nature 2002; 420: 673–678.
Martinek S, Inonog S, Manoukian AS, Young MW : A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 2001; 105: 769–779.
Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J : The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci 2005; 25: 5430–5437.
Ashmore LJ, Sathyanarayanan S, Silvestre DW, Emerson MM, Schotland P, Sehgal A : Novel insights into the regulation of the timeless protein. J Neurosci 2003; 23: 7810–7819.
Shafer OT, Rosbash M, Truman JW : Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci 2002; 22: 5946–5954.
Saez L, Young MW : Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 1996; 17: 911–920.
Glossop NR, Lyons LC, Hardin PE : Interlocked feedback loops within the Drosophila circadian oscillator. Science 1999; 286: 766–768.
Cyran SA, Buchsbaum AM, Reddy KL et al: vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003; 112: 329–341.
Rosato E, Codd V, Mazzotta G et al: Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr Biol 2001; 11: 909–917.
Ceriani MF, Darlington TK, Staknis D et al: Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 1999; 285: 553–556.
Busza A, Emery-Le M, Rosbash M, Emery P : Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science 2004; 304: 1503–1506.
Lin FJ, Song W, Meyer-Bernstein E, Naidoo N, Sehgal A : Photic signaling by cryptochrome in the Drosophila circadian system. Mol Cell Biol 2001; 21: 7287–7294.
Stanewsky R, Kaneko M, Emery P et al: The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 1998; 95: 681–692.
Dissel S, Codd V, Fedic R et al: A constitutively active cryptochrome in Drosophila melanogaster. Nat Neurosci 2004; 7: 834–840.
Emery P, Stanewsky R, Hall JC, Rosbash M : A unique circadian-rhythm photoreceptor. Nature 2000; 404: 456–457.
Yang Z, Emerson M, Su HS, Sehgal A : Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron 1998; 21: 215–223.
Suri V, Qian Z, Hall JC, Rosbash M : Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron 1998; 21: 225–234.
Hunter-Ensor M, Ousley A, Sehgal A : Regulation of the Drosophila protein timeless suggests a mechanism for resetting the circadian clock by light. Cell 1996; 84: 677–685.
Zeng H, Qian Z, Myers MP, Rosbash M : A light-entrainment mechanism for the Drosophila circadian clock. Nature 1996; 380: 129–135.
Plautz JD, Kaneko M, Hall JC, Kay SA : Independent photoreceptive circadian clocks throughout Drosophila. Science 1997; 278: 1632–1635.
Akhtar RA, Reddy AB, Maywood ES et al: Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 2002; 12: 540–550.
Yamazaki S, Numano R, Abe M et al: Resetting central and peripheral circadian oscillators in transgenic rats. Science 2000; 288: 682–685.
Yoo SH, Yamazaki S, Lowrey PL et al: PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 2004; 101: 5339–5346.
Krishnan B, Levine JD, Lynch MK et al: A new role for cryptochrome in a Drosophila circadian oscillator. Nature 2001; 411: 313–317.
Shearman LP, Sriram S, Weaver DR et al: Interacting molecular loops in the mammalian circadian clock. Science 2000; 288: 1013–1019.
Sato TK, Panda S, Miraglia LJ et al: A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004; 43: 527–537.
Preitner N, Damiola F, Lopez-Molina L et al: The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002; 110: 251–260.
Looby P, Loudon AS : Gene duplication and complex circadian clocks in mammals. Trends Genet 2005; 21: 46–53.
Tauber E, Last KS, Olive PJ, Kyriacou CP : Clock gene evolution and functional divergence. J Biol Rhythms 2004; 19: 445–458.
Ohno S : Evolution by Gene Duplication. London, New York: Allen & Unwin, Springer-Verlag, 1970.
Shearman LP, Jin X, Lee C, Reppert SM, Weaver DR : Targeted disruption of the mPer3 gene: subtle effects on circadian clock function. Mol Cell Biol 2000; 20: 6269–6275.
Zylka MJ, Shearman LP, Weaver DR, Reppert SM : Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron 1998; 20: 1103–1110.
Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR : Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 2001; 30: 525–536.
Masubuchi S, Kataoka N, Sassone-Corsi P, Okamura H : Mouse Period1 (mPER1) acts as a circadian adaptor to entrain the oscillator to environmental light/dark cycles by regulating mPER2 protein. J Neurosci 2005; 25: 4719–4724.
Gotter AL, Levine JD, Reppert SM : Sex-linked period genes in the silkmoth, Antheraea pernyi: implications for circadian clock regulation and the evolution of sex chromosomes. Neuron 1999; 24: 953–965.
Benna C, Scannapieco P, Piccin A et al: A second timeless gene in Drosophila shares greater sequence similarity with mammalian tim. Curr Biol 2000; 10: R512–R513.
Gotter AL, Manganaro T, Weaver DR et al: A time-less function for mouse timeless. Nat Neurosci 2000; 3: 755–756.
Barnes JW, Tischkau SA, Barnes JA et al: Requirement of mammalian timeless for circadian rhythmicity. Science 2003; 302: 439–442.
Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A : Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol 2005; 25: 3109–3116.
Krings G, Bastia D : swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA 2004; 101: 14085–14090.
Katou Y, Kanoh Y, Bando M et al: S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003; 424: 1078–1083.
Chan RC, Chan A, Jeon M et al: Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 2003; 424: 1002–1009.
Perry J : Weighing in on a Timeless controversy. Proteins 2005 (in Press).
Kyriacou CP, Odell M : No ARM in it ? Reply to Kippert and Gerloff. Curr Biol 2004; 14: R652–R653.
Vodovar N, Clayton JD, Costa R, Odell M, Kyriacou CP : The Drosophila clock protein Timeless is a member of the Arm/HEAT family. Curr Biol 2002; 12: R610–R611.
Kippert F, Gerloff DL : Timeless and Armadillo: a link too far [Comment on Vodovar et al (2002) Curr Biol 12; R610–R611]. Curr Biol (2004); 14: R650–R651; author reply R652–R653.
Kopelman NM, Lancet D, Yanai I : Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 2005; 37: 588–589.
Collins BH, Rosato E, Kyriacou CP : Seasonal behavior in Drosophila melanogaster requires the photoreceptors, the circadian clock, and phospholipase C. Proc Natl Acad Sci USA 2004; 101: 1945–1950.
Majercak J, Chen WF, Edery I : Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol 2004; 24: 3359–3372.
Majercak J, Sidote D, Hardin PE, Edery I : How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 1999; 24: 219–230.
Kaneko M, Helfrich-Forster C, Hall JC : Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J Neurosci 1997; 17: 6745–6760.
Ewer J, Frisch B, Hamblen-Coyle MJ, Rosbash M, Hall JC : Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. J Neurosci 1992; 12: 3321–3349.
Helfrich-Forster C : Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. J Comp Physiol A 1998; 182: 435–453.
Helfrich-Forster C : The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. Proc Natl Acad Sci USA 1995; 92: 612–616.
Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH : A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 1999; 99: 791–802.
Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC : Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci USA 2000; 97: 3608–3613.
Lin Y, Stormo GD, Taghert PH : The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci 2004; 24: 7951–7957.
Helfrich-Forster C : Neurobiology of the fruit fly's circadian clock. Genes Brain Behav 2005; 4: 65–76.
Helfrich-Forster C : The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc Res Technol 2003; 62: 94–102.
Kaneko M, Hall JC : Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 2000; 422: 66–94.
Helfrich-Forster C, Edwards T, Yasuyama K et al: The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J Neurosci 2002; 22: 9255–9266.
Veleri S, Brandes C, Helfrich-Forster C, Hall JC, Stanewsky R : A self-sustaining, light-entrainable circadian oscillator in the Drosophila brain. Curr Biol 2003; 13: 1758–1767.
Miskiewicz K, Pyza E, Schurmann FW : Ultrastructural characteristics of circadian pacemaker neurones, immunoreactive to an antibody against a pigment-dispersing hormone in the fly's brain. Neurosci Lett 2004; 363: 73–77.
Kaneko M, Park JH, Cheng Y, Hardin PE, Hall JC : Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol 2000; 43: 207–233.
Piggins HD, Antle MC, Rusak B : Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci 1995; 15: 5612–5622.
Piggins HD, Marchant EG, Goguen D, Rusak B : Phase-shifting effects of pituitary adenylate cyclase activating polypeptide on hamster wheel-running rhythms. Neurosci Lett 2001; 305: 25–28.
Shen S, Spratt C, Sheward WJ et al: Overexpression of the human VPAC2 receptor in the suprachiasmatic nucleus alters the circadian phenotype of mice. Proc Natl Acad Sci USA 2000; 97: 11575–11580.
Romijn HJ, Sluiter AA, Pool CW, Wortel J, Buijs RM : Differences in colocalization between Fos and PHI, GRP, VIP and VP in neurons of the rat suprachiasmatic nucleus after a light stimulus during the phase delay versus the phase advance period of the night. J Comp Neurol 1996; 372: 1–8.
Hannibal J : Pituitary adenylate cyclase-activating peptide in the rat central nervous system: an immunohistochemical and in situ hybridization study. J Comp Neurol 2002; 453: 389–417.
Harmar AJ, Marston HM, Shen S et al: The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 2002; 109: 497–508.
Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED : Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 2005; 8: 476–483.
Stoleru D, Peng Y, Agosto J, Rosbash M : Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 2004; 431: 862–868.
Grima B, Chelot E, Xia R, Rouyer F : Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 2004; 431: 869–873.
Lee HS, Billings HJ, Lehman MN : The suprachiasmatic nucleus: a clock of multiple components. J Biol Rhythms 2003; 18: 435–449.
Jagota A, de la Iglesia HO, Schwartz WJ : Morning and evening circadian oscillations in the suprachiasmatic nucleus in vitro. Nat Neurosci 2000; 3: 372–376.
Pittendrigh C, Daan S : A functional analysis of circadian pacemakers in nocturnal rodents. V. Pacemaker structure: a clock for all seasons. J Comp Physiol A 1976; 106: 333–355.
Daan S, Berde C : Two coupled oscillators: simulations of the circadian pacemaker in mammalian activity rhythms. J Theor Biol 1978; 70: 297–313.
Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S : Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 2002; 277: 14048–14052.
Lin Y, Han M, Shimada B et al: Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci USA 2002; 99: 9562–9567.
Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA : Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci 2002; 22: 9305–9319.
Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW : Circadian regulation of gene expression systems in the Drosophila head. Neuron 2001; 32: 657–671.
McDonald MJ, Rosbash M : Microarray analysis and organization of circadian gene expression in Drosophila. Cell 2001; 107: 567–578.
Levine JD, Funes P, Dowse HB, Hall JC : Resetting the circadian clock by social experience in Drosophila melanogaster. Science 2002; 298: 2010–2012.
Stanewsky R, Frisch B, Brandes C, Hamblen-Coyle MJ, Rosbash M, Hall JC : Temporal and spatial expression patterns of transgenes containing increasing amounts of the Drosophila clock gene period and a lacZ reporter: mapping elements of the PER protein involved in circadian cycling. J Neurosci 1997; 17: 676–696.
Acknowledgements
We thank the BBSRC, NERC and Wellcome Trust for supporting our research, and the Royal Society for a Wolfson Research Merit Award to CPK. We also thank three anonymous referees who put up with, and treated with considerable sympathy, a very rough first draft that was written by one of us (CPK) from memory, during a brief stay in an Italian hospital.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rosato, E., Tauber, E. & Kyriacou, C. Molecular genetics of the fruit-fly circadian clock. Eur J Hum Genet 14, 729–738 (2006). https://doi.org/10.1038/sj.ejhg.5201547
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201547
Keywords
This article is cited by
-
Ectopic RING zinc finger gene from hot pepper induces totally different genes in lettuce and tobacco
Molecular Breeding (2018)
-
Circadian Rhythms and Period Expression in the Hawaiian Cricket Genus Laupala
Behavior Genetics (2013)
-
Bumblebee foraging rhythms under the midnight sun measured with radiofrequency identification
BMC Biology (2010)
-
Focus on behavioural genetics
European Journal of Human Genetics (2006)