Abstract
We describe a new member of the human Factor H protein family, termed Factor H-related protein 4A (FHR-4A). The corresponding cDNA sequence was isolated and encodes a secreted protein of 559 amino acids, with a predicted molecular weight of 63.2 kDa. Apparently, this novel cDNA is derived from the human FHR-4 gene. Genetic analysis shows that the human FHR-4 gene is composed of 10 coding exons, and two distinct mRNA transcripts are derived from this gene by alternative splicing. The short FHR-4B form represents a truncated variant and encodes a secreted protein of five domains (previously termed FHR-4). The long transcript encodes the novel FHR-4A protein that is composed of nine complement control protein (CCP) domains. A unique feature of FHR-4A is the tandem arrangement of four CCP domains forming a ‘natural dimer’ of the short isoform. The FHR-4A protein is identified in human plasma as a 86 kDa protein. The difference between the predicted and observed molecular masses is explained by glycosylation. Comparison of the deduced protein sequence of FHR-4A with peptides from a 86 kDa apolipoprotein described by us earlier suggests that the long form, FHR-4A, represents this apoprotein. In summary, FHR-4A is a new Factor H-related protein with a unique domain composition, that is, an internal duplication of four CCP domains. To our knowledge, FHR-4A provides the first evidence for alternative splicing among Factor H-related genes.
Similar content being viewed by others
Introduction
The human Factor H protein family consists of seven related plasma proteins: Factor H, Factor H-like protein-1 (FHL-1) and five Factor H-related proteins (FHR-1, FHR-2, FHR-3, FHR-4 and FHR-5). All members of this protein family share structural and most likely functional similarities. They (i) represent secreted plasma proteins, (ii) are exclusively composed of conserved protein domains termed complement control protein modules (CCPs), (iii) are synthesized primarily by hepatocytes and (iv) are immunologically related to each other and to Factor H.1 The five FHR proteins share sequence similarity of their individual CCP domains, and based on this similarity the proteins can be divided into three distinct subgroups: (i) FHR-1 and FHR-2, (ii) FHR-3 and FHR-4 and (iii) FHR-5.
Factor H and FHL-1 are both derived from the human Factor H gene by alternative splicing.2 The FHL-1 protein represents the first seven CCP domains of Factor H and has four unique amino acids at the C-terminus. All FHR proteins are encoded by separate genes, which similar to the Factor H gene are located in the regulators of complement activation (RCA) gene cluster on the long arm of human chromosome 1 (1q32).3, 4, 5, 6 With one exception each domain of the Factor H family members is encoded by a unique exon, indicating exon shuffling and duplication events as a basis for the diversity of this gene family. The structure of the FHR-4 gene has not been analysed so far.
Factor H and FHL-1 act as important complement regulators, however, the biological function(s) of the FHR proteins is currently unknown. The structural similarity and the sequence identity of individual protein domains suggest overlapping function(s) of FHR proteins and activities shared with the complement and immune regulator Factor H.7 FHRs and Factor H bind the same ligands, as demonstrated for C3b (FHR-1, FHR-3, FHR-4 and FHR-5),8, 9, 10 heparin (FHR-1 and FHR-3, FHR-5),8, 9, 11, 12 C-reactive protein (FHR-3 and FHR-4)13 and some subtypes of the streptococcal M protein (FHR-3).14 FHR-1, FHR-2 and FHR-4 as well as Factor H are present in plasma lipoproteins and FHR-5 has been found in kidney glomerular complement deposits.15 FHR-3 and FHR-4 enhance the cofactor activity of Factor H, most likely by changing the conformation of C3b.9 A relatively weak cofactor activity has been reported for the FHR-5 protein.12 FHR proteins are conserved in evolution and are present in other vertebrates.16, 17, 18, 19, 20 The conservation in evolution suggests an early and important role at least for some FHR proteins.
Recently, a 86 kDa human plasma and apolipoprotein was identified, purified and the sequence of six tryptic peptide fragments was determined.21 The derived peptide sequences showed high similarity but not complete identity to the FHR-4 protein. The recombinant FHR-4 protein has a molecular mass of approximately 42 kDa and, based on the peptide match and the crossreactivity of polyclonal antiserum raised against the 86 kDa lipoprotein, it was hypothesized that FHR-4 forms a homodimer in plasma and represents the 86 kDa lipoprotein. However, differences in sequence and in mobility were observed and explained by polymorphic variations or by different glycosylation patterns of the recombinant versus the native protein. Based on these discrepancies, we hypothesized that the 86 kDa apolipoprotein might be a distinct protein that is highly related to FHR-4. Here we identify a new FHR protein of nine CCP domains, that is identical to the 86 kDa apolipoprotein.
Materials and methods
Library screening
A human liver cDNA library (Stratagene) was screened with a probe representing the FHR-4B cDNA. This probe was labelled with 32P-dCTP (Amersham) using the High Prime DNA Labelling kit (Roche). The library was plated on LB-agar plates and filter lifts (Hybond-N membranes, Amersham) were performed according to standard procedures. The labelled probe was added O/N in 10 × Denhardt's, 1 × HEPES solution containing 100 μg/ml herring sperm DNA (Sigma) at 65°C with shaking. The filters were extensively washed with 2 × SSC containing 0.1% SDS, dried and exposed to Kodak XAR-5 films (Sigma) at –70°C using intensifier screens. Positive clones were isolated, re-screened, and the cDNA inserts were sequenced as recommended.
PCR
For amplification of FHR-4A fragments, a human liver cDNA library or HUH7 cDNA was used. Total RNA was extracted from HUH7 human liver cells and cDNA was synthesized using oligo(dT) primer with the Supercript II RNase H− Reverse Transcriptase kit (Gibco). Specific FHR-4A fragments were amplified using the following primers: (SP) 5′-ATGTTGTTACTAATCAATGTC-3′ for the signal peptide, (4BR2) 5′-CTGGTACTCGACTCTTGACCA-3′ for CCP 4, (R6SCR1F) 5′-CAAGAAGTGAAACCTTGTGAG-3′ for CCP 5, (R6CCP3) 5′-TTGGACTATGAATGCTAT-3′ for CCP 3, (R6CCP6) 5′-AATGCAAATTGGTTGTGC-3′ for CCP 6, (R4SCR5R) 5′-TTCGCATCTGGGGTATTCCAC-3′ for CCP 9 and oligo(dT) primers. The PCR conditions were 33 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, extension at 72°C for 75 s, final extension of 10 min. For the amplification of the full FHR-4A coding sequence, two additional primers were used: (R6F1) 5′-AAGATTTCAAACCCCAAACAG-3′ and (R6R2) 5′-TCAGAAGCTATCTTTGCAAGC-3′ with the Expand Long Template PCR System (Roche) according to the manufacturer's recommendations. Amplified fragments were subcloned into TOPO TA vector (Invitrogen) and the inserts sequenced. The primer pairs and the amplified fragments are shown in Figure 1a.
In silico analysis
The sequences of the clones obtained from the library screening and from the PCR reactions were compared to database sequences via blastn (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov). The individual CCP domains of FHR-4A were aligned using the ClustalW program at the European Bioinformatics Institute (http://www.ebi.ac.uk). The sequence comparisons were performed using the blastp program to compare individual CCP domains to each other. For sequence analysis and handling the GenoDB program was used. The protein sequences used for comparison were from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) protein sequence databank, Factor H (CAA68704), FHR-1 (CAA39666), FHR-2 (CAA60375), FHR-3 (CAA48639), FHR-4 (CAA66980), FHR-5 (AAK15619) and the mouse FHR protein (sequence originally compiled from clones 3A4 and 5G4, AAA37415). The nucleotide sequences were FHR-4 (X98337) and the mouse FHR protein 3A4/5G4 (M29010).
The molecular mass and pI value for FHR-4A were calculated using the relevant programs at the Expert Protein Analysis System (ExPASy) proteomics server of the Swiss Institute of Bioinformatics (http://us.expasy.org/).
For genomic analysis three contigs were used from the Human Genome database (http://www.ncbi.nih.gov/Genomes). The chromosome 1 fragment BX248415 contains the N-terminal part of FHR-4A (exons coding for signal peptide, CCPs 1–5). The chromosome 1 sequence AL139418 begins with the C-terminal part of the FHR-4 gene (exons coding for CCPs 5–9 and 3′ UTR) and overlaps with the previous sequence. The genomic contig NT_004671 was used to confirm the FHR-4A sequence and to analyse the exon–intron structure of FHR-4A. The chimpanzee chromosome 1 sequence was downloaded from http://genome.ucsc.edu.
Western blotting
Human plasma (1 μl) was separated by SDS-PAGE under nonreducing conditions and transferred to nitrocellulose membrane. The membrane was blocked in 3% skim milk and developed using rabbit polyclonal FHR-4B antiserum (1:1000 dilution)21 or Factor H-specific goat antiserum (Calbiochem) with the corresponding HRP-conjugated secondary antibodies (DAKO, diluted 1:1000) by the addition of 0.3% (wt/vol) 4-chloro-1-naphtol (Sigma-Aldrich) as chromogen. To study the N-attached glycan chains in FHR-4A, human plasma was incubated with 1 U of N-glycosidase F (Roche) overnight at 37°C, then blotted and developed with anti-FHR-4B antibody as described above.
Results
Identification of a Factor H-related cDNA
In order to identify new Factor H-related protein(s), a human liver cDNA library was screened with a probe representing FHR-4B. One clone termed 8/I was isolated and sequenced. The sequence of this clone was highly similar to, but clearly distinct from the 5′ region of the FHR-4B cDNA. The remaining part of the novel cDNA was amplified by specific primers (Figure 1a). In addition, a full coding sequence was amplified using primers specific to the 5′ (primer R6F1) and 3′ (primer R6R2) untranslated regions. The full-length cDNA sequence is 2030 nucleotides long (Figure 1b). It includes a translational initiation site (TCT AAC ATG) at positions 61–69 and a poly(A) signal site (AATAAA) at positions 2007–2012. Two overlapping genomic fragments (BX248415 and AL139418) include this cDNA sequence.
The FHR-4A protein
The nucleotide sequence encodes an open reading frame of 578 amino acids. The deduced protein sequence includes an N-terminal hydrophobic signal peptide that is identical to the signal peptide of FHR-4B (residues 1–19), thus indicating that the encoded protein is expressed via the secretory pathway. The predicted molecular mass of the secreted nonglycosylated protein is 63.2 kDa, with a predicted pI value of 4.85. The amino-acid sequence includes seven consensus sites for N-linked glycosylation (NXS/T) at positions 127–129 (NSS), 186–188 (NTT), 206–208 (NSS), 374–376 (NSS), 433–435 (NTT), 453–455 (NSS) and 557–559 (NTS).
The secreted protein has a length of 559 amino acids and is exclusively composed of nine CCP domains. Each domain has the four essential Cys residues and includes additional conserved amino acids (Figure 2a). Homology comparison of the individual domains to each other reveals an internal duplication of four CCP domains (Figure 2b). Domains 1–4 (repeat A) display striking similarity to domains 5–8 (repeat B). These CCP domains show a strong similarity to other FHR proteins, particularly to FHR-4B and to Factor H. Thus, we conclude that the sequence represents a novel FHR protein, which, based on the strong sequence similarity and genetic analysis (see below) is termed FHR-4A. On the amino-acid level, CCP domain 1 of FHR-4A shows 98% identity to domain 1 of FHR-4B, these domains differ in five nucleotides and one single amino acid. CCP domains 6–9 of FHR-4A are identical to domains 2–5 of the FHR-4B protein. The individual domains of the repetitive motif of FHR-4A protein show the strongest homology to domains 6, 8, 9 and 19 of Factor H. Domains 8 and 9 of FHR-4A show strong similarity (i.e 63 and 39% identity) to the two most C-terminal domains of Factor H, that is, CCP domains 19 and 20 (Table 1).
Structure of the FHR-4A protein. (a) The secreted FHR-4A protein is composed of nine consecutive CCP domains. The conserved Cys residues are shown inverted and potential N-linked glycosylation sites are underlined. (b) The FHR-4A protein contains two homologous repeat regions (A, B), as indicated by the arrows.
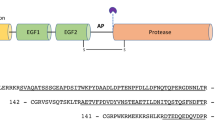
These analyses indicate that FHR-4A is highly related to FHR-4B. Therefore, we examined whether FHR-4A might represent the 86 kDa plasma apolipoprotein that was originally isolated from human chylomicrons.21 Six tryptic peptide fragments of the 86 kDa plasma apolipoprotein21 show a complete match with the sequence of the predicted FHR-4A protein (Figure 3). Peptide I spans from position 265 to position 286 linking CCP 4 (homologue of domain 19 of Factor H) to CCP 5 (homologue of domain 6 of Factor H). Peptide II (positions 156–168 and 403–415) and peptide III (positions 175–181 and 422–428) match to both CCP 3 and CCP 7 of FHR-4A. Peptide IV matches positions 182–192, peptide V to positions 229–248 and peptide VI to the C-terminus of the FHR-4A protein, positions 568–578. Based on the identities, we conclude that the FHR-4A protein represents the 86 kDa apolipoprotein.
Sequence comparison of FHR-4A domains with the related domains of Factor H and FHR proteins. (a) The nine CCP domains of FHR-4A (FHR-4A/1–FHR-4A/9) are aligned with the most related CCP domains of other FHR proteins and Factor H. Identical residues are indicated as dots. Above the CCP domains six peptide sequences (labelled with Roman numbers) are shown that are derived from the 86 kDa plasma apolipoprotein.21 The amino acids in these peptides that match specifically to the FHR-4A sequence are shown inverted. ‘X’ indicates an unidentified residue. Note that peptide I spans CCP 4 and CCP 5. (b) Vertical alignment of the related domains of FHR-4A, FHR-4B and Factor H. The domains of the individual proteins are numbered consecutively and are aligned to the most similar domain in Factor H/FHR-4B. The CCP domains of FHR-4A that are identical to domains of FHR-4B are shown in black.
Expression of FHR-4A in human plasma
Expression of the FHR-4A protein in human plasma was analysed by SDS-PAGE and Western blotting, using a polyclonal antiserum raised against recombinant FHR-4B. Under nonreducing conditions a band of 86 kDa representing FHR-4A was identified (Figure 4a, lane 1). The smaller band of ca. 45 kDa most likely represents the monomeric native FHR-4B protein. To prove glycosylation of the FHR-4A protein and to explain the difference between the observed and the calculated molecular masses, a serum sample was incubated with N-glycosidase F (PNGase). Upon PNGase treatment the mobility of FHR-4A was increased and the protein appeared as a doublet (Figure 4a, lane 2). FHR-4A is the dominant FHR-4 isoform in plasma (Figure 4b, lane 1). As a control, a Factor H-specific antiserum that identified the 150 kDa Factor H protein, FHL-1 and the two forms of FHR-1 was used (Figure 4b, lane 2). This antiserum reacted weakly with both FHR-4A and FHR-4B. Levels of FHR-4A and FHR-4B vary in plasma derived from different individuals (Figure 4c).
FHR-4A is a plasma glycoprotein. (a) Human plasma treated with (lane 2) or without (lane 1) N-Glycosidase F (PNGase) was separated by SDS-PAGE under nonreducing conditions, followed by Western blotting and developed with an antiserum raised against recombinant FHR-4B. (b) Western blot of human plasma developed with anti-FHR-4B (lane 1) or anti-Factor H (lane 2) antibodies shows that FHR-4A is the prominent plasma protein. (c) Variation of FHR-4A levels in plasma of different individuals (lanes 1–8). The blot was developed as above. The mobility of the markers is indicated on the left. Albumin affects the mobility of the proteins in the 45–60 kDa range, therefore distinguishing between FHR-4B (ca. 45 kDa) and the cross-reacting FHR-3 protein (multiple bands of ca. 45–56 kDa) is not possible.
The human FHR-4 gene: in silico genomic analysis
The FHR-4 gene is contained in a contig derived from human chromosome 1 (NT_004671, GI 29793399). The FHR-4 gene is positioned within the RCA gene cluster between the FHR-1 and FHR-2 genes (Figure 5a). The gene is organized in 10 exons (shown in black in Figure 5), representing the signal peptide and the nine CCP domains that all show a phase 1–1 organization (Table 2). Apparently, FHR-4A and FHR-4B share six exons and are derived from the same gene by alternative splicing. The one amino-acid difference in CCP 1 might be due to polymorphism as in the genome no additional exon variant or related gene can be found. Exon II was sequenced from 10 individuals and the sequences were identical (data not shown).
The human FHR-4 gene. (a) The human regulators of complement activation (RCA) gene cluster is shown schematically (based on the data from the human genome sequencing, build 23 at http://www.ncbi.nlm.nih.gov). The Factor H family members represent one gene cluster which is separated from the other RCA genes, in good accordance with their evolutionary relationship.16 The FHR-4 gene is shown in black. The structure of the gene is shown below (drawn to scale). The FHR-4 gene is organized in 10 exons coding for the signal peptide and nine CCP domains, which are shown in black. (b) Schematic representation of the FHR-4 gene. The coding exons are shown in black and are indicated by Roman numbers, while non-coding exons are white and labelled with letters a, b and c. The two repeat regions (A, B) are shown by the arrows above the exons. A human EST sequence having different exon combination is also shown. The mouse FHR cDNA clone (3A4/5G4) shows similar exon arrangement to that of the FHR-4 gene. Exons coding for CCPs related to Factor H domains 6–9 and 19 (internal homologous repeat) are duplicated in FHR-4 but not in the mouse FHR protein. (c) Alignment of FHR-4A domains from human (hs) and chimpanzee (pt). Identical residues are shown as dots. Due to gaps in the available chimpanzee genomic sequence, domain 4 is not complete and CCPs 7 and 8 are missing.
A detailed analysis of the intronic regions revealed between exon I and exon II a sequence homologous to CCP domain 5 of Factor H. Similarly, within the introns spanning exon II and exon III and between exon VI and exon VII sequences homologous to CCP domain 7 of Factor H were identified. However, these three exons are noncoding since on the protein level they each lack one of the essential Cys residues characteristic to CCP domains and necessary for proper disulphide bridge formation and folding. These sequences are indicative of the duplication event which gave rise to the FHR-4 gene. The sequences related to CCP 5 and 7 of Factor H are also identified in the human EST clone Hs1 4828 33 43 1 (Figure 5b).
The FHR-4 gene is conserved in evolution. The CCP composition of a mouse FHR protein identified on the cDNA level (clone 3A4/5G4)17 shows a rather similar domain structure as the human FHR-4 gene. The mouse transcript includes the two CCP-related sequences that are found in FHR-4 introns, but represents a monomeric form prior to the internal duplication event (Figure 5b). In addition, CCP sequences almost identical to the CCP domains 1–3, 5, 6 and 9 of FHR-4A are found in the chimpanzee genome (Figure 5c).
Discussion
In this report, we show that the human FHR-4 gene codes for at least two proteins. The long isoform described here is a novel member of the human Factor H protein family and is termed FHR-4A. The previously described FHR-4 protein represents the short isoform and is now termed FHR-4B. The FHR-4A cDNA is expressed in the liver and the protein is composed of nine CCP domains (Figures 1 and 2). The individual domains show high similarity to each other and to other FHR proteins (Table 1 and Figure 3). The internal similarity of the FHR-4A CCP domains reveals a duplication of two consecutive motifs, each of four CCP domains in length (Figure 3). This is the first example of a tandem repeat motif in a Factor H-related protein.
The FHR-4A protein is a plasma glycoprotein as confirmed by Western blot analysis (Figure 4). Six tryptic peptide fragments (described earlier21) derived from a 86 kDa apolipoprotein show a perfect match with the predicted FHR-4A protein sequence (Figure 3), confirming the existence of FHR-4A. The perfect match of these peptides with the FHR-4A protein and the mismatches with the FHR-4B protein indicate that the FHR-4A cDNA encodes the 86 kDa apoprotein (Figures 3 and 4 and Skerka et al21). In addition, the apoprotein appeared as a 63 kDa band upon extensive reduction,21 in good agreement with the predicted molecular weight of nonglycosylated FHR-4A (63.2 kDa) and with the size of PGNase-treated plasma FHR-4A (Figure 4).
FHR-4A is considered a natural dimer of the FHR-4B protein. It is therefore expected that FHR-4A shares the binding characteristics of FHR-4B (binding to C3b and to C-reactive protein)9, 13, 22 and functions. Due to the domain duplications, FHR-4A has probably double binding sites for at least some ligands and most likely binds these ligands with higher avidity and, consequently, may display enhanced biological activities. Similar to FHR-4B, also FHR-4A may enhance Factor H-mediated cofactor activity.9
The human FHR-4 gene was analysed using a bioinformatics approach. This gene has 10 exons (Figure 5 and Table 2). Both FHR-4A and FHR-4B are encoded by the FHR-4 gene and their transcripts are derived by means of alternative splicing. The FHR-4A and the FHR-4B cDNAs share six exons (I, II, VII–X) and the protein products share CCP domains 1 and 6–9 (shown in black in Figure 3). Exon II was sequenced from genomic DNA of 10 individuals and no sequence variations were observed (data not shown). In the human genome sequence only this exon is identified by database search, confirming the model of alternative splicing. Therefore, the five nucleotide differences in CCP 1 of FHR-4A compared to the previously reported FHR-4B cDNA sequence are likely due to polymorphism.
Within three introns of the FHR-4 gene, sequences that are homologous to CCP 5 and CCP 7 of factor H are present and are indicative of a duplication event (Figure 5). These sequences show relatively weak similarity (ca. 40%) to the corresponding sequences in Factor H. Using specific primers incorrectly spliced transcripts of the FHR-4 gene that contain these CCP-related sequences and intronic sequences of various lengths have been amplified from liver cDNA (data not shown). Likewise, in EST database a sequence that includes CCP 5- and CCP 7-homologue sequences is present (Figure 5b). A splice variant of FHR-4A which lacks the first 24 nucleotides of CCP 1 was also amplified by PCR. This eight amino acid-long deletion is due to the presence of a cryptic splicing site in CCP 1 (gtg aaa cct tgt gat ttt cca gaa att).
The human FHR-4 gene is highly related in structure and sequence to a mouse protein encoded by two cDNA clones named 3A4 and 5G4 (Figure 5b).17 This mouse cDNA codes for domains homologous to CCP domains 5–9 and 19–20 of Factor H. The signal peptide of human FHR-4 is more similar to the signal peptide of this mouse FHR protein (58% identity) than to the signal peptide of mouse or human Factor H (both 42%). The structural and sequence similarities between the mouse and the human FHR proteins indicate a conservation and evolution independent of Factor H. In addition, a homologue of the FHR-4 gene is identified in the chimpanzee genome (Figure 5c).
Expression of the FHR-4A mRNA was previously shown by Northern blot analysis of human liver mRNA.21 An FHR-4B fragment hybridized to mRNA species of 1.4, 2.2 and 3.5 kb, demonstrating expression of three distinct FHR-4-related transcripts.21 The 1.4 kb mRNA represents FHR-4B, and based on the length of the FHR-4A cDNA it is suggested that the 2.2 kb species corresponds to FHR-4A. The nature of the 3.5 kb species is yet unknown.
In summary, by combining experimental data with data mining, we identified and characterized FHR-4A, a novel member of the Factor H protein family. In addition, the structure of the FHR-4 gene was analysed and shown to code for two FHR-4 isoforms, providing the first evidence for alternative splicing among the FHR genes.
References
Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S : The Factor H protein family. Immunopharmacology 1999; 42: 53–60.
Schwaeble W, Zwirner J, Schulz TF, Linke RP, Dierich MP, Weiss EH : Human complement factor H: expression of an additional truncated gene product of 43 kDa in human liver. Eur J Immunol 1987; 17: 1485–1489.
Skerka C, Moulds JM, Taillon-Miller P, Hourcade D, Zipfel PF : The human Factor H-related gene 2 (FHR2): structure and linkage to the coagulation Factor XIIIb gene. Immunogenetics 1995; 42: 268–274.
Rodríguez de Córdoba S, Díaz-Guillén MA, Heine-Suñer D : An integrated map of the human regulator of complement activation (RCA) gene cluster on 1q32. Mol Immunol 1999; 36: 803–808.
Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL : Complement Factor H: sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 2000; 37: 41–52.
McRae JL, Murphy BE, Eyre HJ, Sutherland GR, Crawford J, Cowan PJ : Location and structure of the human FHR-5 gene. Genetica 2002; 114: 157–161.
Zipfel PF, Skerka C : Complement Factor H and related proteins: an expanding family of complement-regulatory proteins? Immunol Today 1994; 15: 121–126.
Majno SA, Jokiranta TS, Hühn M, Seeberger H, Hellwage J : The human Factor H-related protein 1 (FHR-1) binds to C3b and heparin: implications for a regulatory role on the level of C3. Mol Immunol 2003; 40: 174, (abstract).
Hellwage J, Jokiranta TS, Koistinen V, Vaarala O, Meri S, Zipfel PF : Functional properties of complement Factor H-related proteins FHR-3 and FHR-4: binding to the C3d region of C3b and differential regulation by heparin. FEBS Lett 1999; 462: 345–352.
McRae JL, Cowan PJ, Power DA et al: Human Factor H-related protein 5 (FHR-5). A new complement-associated protein. J Biol Chem 2001; 276: 6747–6754.
Prodinger WM, Hellwage J, Spruth M, Dierich MP, Zipfel PF : The C-terminus of Factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to Factor H and Factor H-related proteins. Biochem J 1998; 331: 41–47.
Duthy TG, McRae JL, Griggs KM, Murphy BF, Gordon DL : Functional characterization of human Factor H-related protein 5 (FHR-5) reveals heparin binding and novel cofactor activity. Int Immunopharmacol 2002; 2: 1269, (abstract).
Hellwage J, Jarva H, Zipfel PF, Meri S : Factor H-related proteins interact with C-reactive protein. Immunopharmacology 2000; 49: 70, (abstract).
Kotarsky H, Hellwage J, Johnsson E et al: Identification of a domain in human Factor H and Factor H-like protein-1 required for the interaction with streptococcal M proteins. J Immunol 1998; 160: 3349–3354.
Murphy B, Georgiou T, Machet D, Hill P, McRae J : Factor H-related protein-5: a novel component of human glomerular immune deposits. Am J Kidney Dis 2002; 39: 24–27.
Krushkal J, Bat O, Gigli I : Evolutionary relationships among proteins encoded by the regulator of complement activation gene cluster. Mol Biol Evol 2000; 17: 1718–1730.
Vik DP, Munoz-Canoves P, Kozono H, Martin LG, Tack BF, Chaplin DD : Identification and sequence analysis of four complement Factor H-related transcripts in mouse liver. J Biol Chem 1990; 265: 3193–3201.
Ren G, Doshi M, Hack BK, Alexander JJ, Quigg RJ : Isolation and characterization of a novel rat Factor H-related protein that is up-regulated in glomeruli under complement attack. J Biol Chem 2002; 277: 48351–48358.
Dahmen A, Kaidoh T, Zipfel PF, Gigli I : Cloning and characterization of a cDNA representing a putative complement-regulatory plasma protein from barred sand bass (Parablax neblifer). Biochem J 1994; 301: 391–397.
Zipfel PF, Kemper C, Dahmen A, Gigli I : Cloning and recombinant expression of a barred sand bass (Paralabrax nebulifer) cDNA. The encoded protein displays structural homology and immunological crossreactivity to human complement/cofactor related plasma proteins. Dev Comp Immunol 1996; 20: 407–416.
Skerka C, Hellwage J, Weber W et al: The human Factor H-related protein 4 (FHR-4). A novel short consensus repeat-containing protein is associated with human triglyceride-rich lipoproteins. J Biol Chem 1997; 272: 5627–5634.
Hellwage J, Skerka C, Zipfel PF : Biochemical and functional characterization of the factor-H-related protein 4 (FHR-4). Immunopharmacology 1997; 38: 149–157.
Acknowledgements
We thank Monika Nguyen and Dr Raimund Eck for help with the library screening. This work was supported by the Deutsche Forschungsgemeinschaft, the Thüringer Ministerium für Wissenschaft, Forschung und Kunst and the Kidneeds Foundation, Cedar Rapids, USA. MJ was supported by the Joseph Eötvös Scholarship Public Foundation, Budapest, Hungary and by the Alexander von Humboldt Foundation, Bonn, Germany.
Author information
Authors and Affiliations
Corresponding author
Additional information
Data presented in this paper strongly suggest that the previously described FHR-4 protein is the short isoform deriving from the FHR-4 gene. To avoid confusion we suggest that this protein should be termed FHR-4B, and the newly described long isoform FHR-4A. The nucleotide sequence of FHR-4A is available in the EMBL database under accession number AJ640130.
Rights and permissions
About this article
Cite this article
Józsi, M., Richter, H., Löschmann, I. et al. FHR-4A: a new factor H-related protein is encoded by the human FHR-4 gene. Eur J Hum Genet 13, 321–329 (2005). https://doi.org/10.1038/sj.ejhg.5201324
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5201324
Keywords
This article is cited by
-
Successful therapy of C3Nef-positive C3 glomerulopathy with plasma therapy and immunosuppression
Pediatric Nephrology (2015)
-
DEAP-HUS: Deficiency of CFHR plasma proteins and autoantibody-positive form of hemolytic uremic syndrome
Pediatric Nephrology (2010)
-
A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration
Nature Genetics (2006)
-
Two factor H-related proteins from the mouse: expression analysis and functional characterization
Immunogenetics (2006)