Abstract
We report the molecular characterization of a patient with mild craniofacial and acallosal central nervous system midline defects and a t(4;14)(q25)(q13) chromosome. With the use of flow sorted chromosomes, the translocation breakpoint junction was defined within a 100 kb region with markers mapping to chromosomes 4q25 and 14q13. Analysis of genomic sequences demonstrated that the breakpoint junction at 14q13 was within the third intron of the 5′ untranslated region of the MIPOL1 gene (GI: 22048098). On chromosome 4q25, two breakpoint junctions were found. One was about 47 kb distal to the 5′ end of a putative gene (GI: 8923996) with unknown function but with partial similarity to kinases, and a second breakpoint was within the 3′ end of the PITX2 gene (GI: 21361182) that resulted in the deletion of exons 6 and 7 of this gene. We also searched for microdeletions in a panel of candidate genes mapping within 2Mb of the translocation breakpoint junction on chromosomes 4 and 14, however, no evidence for deletions or rearrangements was found. The finding of two breaks on chromosome 4q25 suggests a complex microrearrangement, such as an inversion, in addition to a translocation in this patient.
Similar content being viewed by others
Introduction
Translocation chromosomes involving human chromosomes 4 and 14 are relatively rare. No previous cases have been reported involving translocations between chromosomes 4q25 and 14q13. Only a few cases of translocations involving chromosomes 4q25 or 14q13 with other chromosomes have been reported.1,2,3,4,5,6 A number of disease genes have been identified thus far at 4q25, including IF (I factor) of C3b inactivator deficiency and PITX2 (paired-like homeodomain transcription factor 2) for Rieger syndrome; and at 14q13, PAX9 (paired box 9) for oligodontia, MIPOL1 (mirror image polydactyly 1) for tetramelic mirror image polydactyly and COCH (cochlin) for dominant deafness 9 (www3.ncbi.nlm.nih.gov/omim, OMIM accession numbers 217030, 601542, 167416, 603196; Kondoh et al6). These cytogenetic intervals also contain genes for various forms of malignant tumors (reviewed by Kamnasaran and Cox,7 cgap.nci.nih.gov/Chromosomes/ Mitelman). The use of positional cloning, including linkage and mutation studies, has primarily defined and identified these disease loci and genes. However, numerous diseases including those mapping to 4q25 and 14q13 were identified using chromosome aberrations. Furthermore, chromosome aberrations have contributed to defining novel phenotypes whereby a single or small number of genes are concurrently affected with respect to altered gene regulation or protein coding integrity.
The PITX2 gene is a member of the bicoid-like homeobox family of transcription factors. The gene, about 20 kb in size, has seven exons that encode four alternative transcripts (PITX2 A,B,C,D).8 Each isoform is made by alternative splicing and differential use of three promoters. Two internal promoters reside in the third and fourth introns, respectively. The protein sequences encoded by all isoforms share in common the homeodomain (exon 6) and C-terminal domain (exon 7). The C-terminal region contains the aristaless domain necessary for mediating protein-protein interactions.8 All isoforms contain dis-similar N-terminal domains, which are speculated to be required for the interaction of the PITX2 protein with other transcription factors.8 The PITX2B protein forms heterodimers with either PITX2A or PITX2C.6 One isoform (PITX2D), that has a truncated homeodomain and complete C-terminal domain, negatively regulates PITX2 transcriptional activity by forming heterodimers with one of the three other isoforms and perhaps prevents interaction of the heterodimer with other transcription factors.8 Studies predominantly in chick and mouse have suggested that the PITX2 isoforms act independently or in overlapping domains with different doses to pattern brain, tooth, heart, lung and gut development.9,10,11 Recent studies have suggested other roles of the PITX2 gene isoforms in hematopoietic stem cell differentiation,12 acute lymphocytic leukemia13 and actin-myosin reorganization.14
The PITX2 gene was initially identified as the gene for Axenfeld–Rieger syndrome type 1 based on the finding of mutations predominantly within the homeodomain.15 Other studies have also shown the PITX2 gene mutated in patients with Peter anomaly,16 iris hypoplasia and iridogoniodysgenesis type 3.17 Axenfield–Rieger syndrome type 1 (ARS1) is an autosomal dominant disorder, in which patients are affected with hypoplasia of the midface, teeth and a range of ocular defects. Other defects of the limb, heart, pituitary, genital and umbilical skin are also prevalent to varying extents among ARS1 patients. Axenfeld–Rieger syndrome type 1 at 4q25 has also been described in a translocation patient with acallosal defects.4
The MIPOL1 gene contains 15 exons that span about 350 kb.6 This gene encodes a 442 amino-acid protein that has two coiled-coil domains in the C terminus, one of which is highly similar to the caspase recruitment domain. The first four exons of the gene are non-coding. The gene has three alternative transcripts of 7, 3 and 2 kb sizes and is expressed in adult kidney, skeletal muscles, liver, heart, pancreas and fetal kidney. Mouse embryonic expression studies show ubiquitous expression of the murine Mipol1 gene during E10.5–E13.5 stages.6 The 10th intron, the MIPOL1 gene has been previously reported disrupted in a t(2;14)(p23.3q13) patient affected with tetramelic mirror image polydactyly and left inguinal hernia.6
In this report, we characterized the translocation breakpoint junction on chromosomes 4 and 14 in a patient with mild craniofacial and acallosal central nervous system midline defects. The genomic sequence spanning the breakpoint junction on both chromosomes was analyzed for disrupted genes. We found a disruption of the MIPOL1 gene at 14q13 and the PITX2 gene at 4q25.
Patient description
The proband was a white male, born at 41 weeks gestation following an uncomplicated pregnancy to a 21-year-old G1 mother. His birth weight was 8 lbs 3 oz and microcephaly was noted at birth. At 7 months, his head circumference was 40.4 cm (50th percentile for 4 months) and length was 63.8 (40th percentile for 4 months). He had a flat face, slightly low set ears, bilateral epicanthal folds, normal interpupillary distance, high palate and normal dermatoglyphics. His limbs were normal. In addition, he demonstrated no ocular anomalies at the age of 7 months. His tone was increased with arching when held. He had feeding and weight gain problems that required feeding with a gastrostomy tube. At 13 months, he could not yet sit or walk and had poor head control. He was able to smile, but did not follow. His height was 50th percentile for 5 months and head circumference was 50th percentile for 4 months. At 19 months, his length was 50th percentile for 11 months and head circumference was 50th percentile for 5 months. Furthermore, he did not exhibit any known dental anomalies at this age. Cranial imaging showed agenesis of the corpus callosum. The patient died during his early childhood. Both parents are clinically normal and have normal karyotypes. The proband's karyotype was ascertained as 46,XY,t(4;14)(q25;q13) based on standard GTG metaphase chromosomes from skin fibroblasts and peripheral blood lymphocyte cultures. No complex rearrangement, other than a translocation, was apparent from his karyotype.
Methods and materials
Flow sorting aberrant chromosomes and breakpoint mapping
A total of 300–500 of the aberrant sorted chromosomes were flow sorted from a lymphoblast cell line and subjected to DOP-PCR amplification before used as template for mapping.1,18 In all, six chromosome 14q13–q21 and 14 chromosome 4q25 specific markers were selected from Ensemble (www.ensembl.org) and the Whitehead Institute CEPH STS content YAC maps (www-genome.wi.mit.edu) for mapping. An additional 12 amplimers were selected from Matsumoto et al19 for more precise mapping at 14q13. Four markers 3′ of the PITX2 gene at 4q25, namely R380DK1, R380DK2, R380DK3 and R380DK4 were designed from the genomic sequence of BAC clone R380D23 (GI:18042428), using primer premiere 3 (www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). Three of each DOP-PCR-amplified derivative chromosomes were pooled before being used for PCR analyses, in 20 μl reactions containing either 40 ng genomic DNA, or 2 μl of pooled DOP-PCR-amplified flow-sorted chromosomes. The PCR conditions and cycles used for each marker are reported by Research Genetics (Huntsville, AL, USA), Matsumoto et al19 and Table 1.
Analysis of the genomic sequence for genes at 4q25 and 14q13
Annotation of the genomic sequence was retrieved from Ensemble (www.ensembl.org) and the National Centre for Bioinformatics Institute MapViewer (www.ncbi.nlm.nih.gov/genome/guide/human) to identify genes and to estimate physical map distances. BLAST searches (www.ncbi.nlm.nih.gov/BLAST) were done on selected genomic BAC clone sequences to precisely map and estimate physical distances at the translocation breakpoint junction.
Analysis of candidate genes for microrearrangement using flow-sorted chromosomes
Amplimers for the MIPOL1, NKX2.1, NKX2.9, SLC25A21, PITX2, PRO0971, T2BP and ANK2 genes were reported previously (UniSTS, www.ncbi.nlm.nih.gov) or designed with Primer Primere 3 (www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). Three of each DOP-PCR-amplified derivative chromosomes were pooled before used for PCR analyses as mentioned above. The PCR primers, conditions and cycles are reported in Table 1.
Analysis of the PITX2 gene for microrearrangement by densitometric analysis
HindIII digested lymphoblast genomic DNA from four normal subjects and the proband was electrophoresed, subjected to Southern transfer onto Hybond N+ filters (Amersham Pharmacia), and hybridized with ExpressHyb solution as specified by ClonTech. Co-hybridizations were done with a single copy 599 bp control probe and either a 432 bp PITX2 5′ or 499 bp PITX2 3′ probe. The templates for the probes were made from PCR products using primers, conditions and cycles listed in Table 1. All probes were radiolabelled with α-32P-dCTP (10 mCi/ml) using the Rediprime kit (Amersham Pharmacia). Washed blots were wrapped in Saran and exposed in Phosporimager cassettes for up to 5 days. The phosphorimager screens were scanned with a Typhoon 8600 variable mode imager (Molecular Dynamics) and densitometric analyses were performed with the ImageQuant version 5.2 software (Molecular Dynamics). The ratio of signal intensity (counts per minute) was calculated in each lane between the control and PITX2 5′ or 3′ bands. All results were within the range of linearity of the instrument.
Results
Defining the breakpoint junction
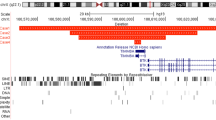
The translocation breakpoint junction was defined between the markers D4S2449 and Q9H1 on chromosome 4 using flow-sorted chromosome analysis (Figure 1). Both markers resided within an estimated physical distance of 45 kb. Furthermore, both D4S2449 and Q9H1 markers resided on BAC clone R477G18 (GI: 17136167). On chromosome 14, the translocation breakpoint junction was initially defined between the markers AFM200ZH4 and D14S306, an estimated 750 kb interval. However, by using a panel of amplimers reported by Matsumoto et al,19 the breakpoint junction was defined more precisely between the markers B319SP6 and B368T7, an estimated 3.9 kb apart, and localized on BAC clone R158A24 (GI:9967624). No deletions were found for any of the chromosomes 4 and 14 markers tested, except for the marker R380DK4 that mapped 25 kb telomeric to the 3′ end of the PITX2 gene (Figure 1).
Fine breakpoint mapping of patient DNA on (a) chromosome 14 and (b) chromosome 4, using flow-sorted chromosomes. Dashed lines=translocation breakpoint junction; markers in italics are reported by Matsumoto et al;19 Candidate genes analyzed are in bold. Not drawn to scale.
Analyses of candidate genes for microrearrangements
Publicly available annotations of the human genomic sequence spanning the translocation breakpoint junctions on chromosomes 4q25 and 14q13 were examined for candidate genes, to search for microrearrangements. Candidate genes were selected based on mapping close to the breakpoint junction and/or having an associated function in the development of the central nervous system. Analysis of the breakpoint junction on chromosome 14q13 showed that the markers B368T7 and B319SP6 mapped within the third and fifth introns respectively of the MIPOL1 gene6 (Figure 2). In order to precisely define the distal limits of the breakpoint junction in the MIPOL1 gene, physical mapping was done with an additional two markers, namely MPDK1 and MPDK2, which were designed within the fourth and fifth exons of this gene. The subsequent mapping with these markers, showed that the breakpoint junction was between B368T7 and MPDK1, which is in the third intron and in the 5′ untranslated region (UTR) of the MIPOL1 gene. An examination of the genomic sequence at the breakpoint junction on chromosome 14q13 revealed that the 5′ end of the SLC25A21 gene (GI:13449278) was the closest gene, mapping within 71 kb proximal to the breakpoint. The SLC25A21 gene (about 133 kb in size) encodes an oxodicarboxylate transport protein, as part of a family of genes involved in mitochondrial metabolism.20 NKX2.1 (GI:4507714), about 3.8 kb in size and NKX2.9 (GI:7657378), about 2 kb in size, physically mapped about 366 and 302 kb, respectively, proximal to the breakpoint junction at 14q13. A homozygous knockout of the murine Nkx2.1 gene showed concepti with defective patterning of the ventral base of the diencephalon and other extracranial anomalies.21 Furthermore, the concepti had partial midline septal fusion defects of the central nervous system with absent structures including the mamillary bodies and pituitary. The murine Nkx2.9 has prominent expression in domains of the developing neural tube and ventral brain and is possibly involved in the patterning of a subset of neurons along the neuroaxis, in particular those of the ventral domains of the developing brain.22 No murine knockout model is yet reported for Nkx2.9. Using a panel of amplimers designed from the 5′ and 3′ ends of SLC25A21, NKX2.1 and NKX2.9 of 14q13, no deletions or rearrangements were detected (Figures 1 and 3).
Analyses of the genomic sequence of 4q25 demonstrated that the 5′ end of the PROO971 gene (about 38 kb in size), a putative gene with unknown function but partial similarity to kinases (GI:8923996), was the closest gene physically mapping within 47 kb distal to the breakpoint junction between D4S2449 and Q9H1. The next closest gene mapping about 91 kb distal to this breakpoint junction is T2BP (GI: 17436598), about 9.8 kb in size. This gene has an unknown function and identity. The ANK2 gene (GI: 21756453), another possible candidate, mapped at least 950 kb distal to this breakpoint junction. The ANK2 gene, about 333 kb in size with three transcript isoforms, belongs to a family of integral membrane proteins involved in diverse roles such as cell motility, activation, proliferation and cell–cell contact. Studies in rat and Drosophila previously showed that the ANK2 ortholog had predominant expression in embryonic, neonatal and adult brain, particularly in areas rich in axons and dendrites.23,24 The Ank2 protein is selectively targeted to premyelinated axons, and is possibly involved in neuronal migration. Using a panel of amplimers designed within the PRO0971, T2BP and ANK2 genes, no deletions or rearrangements among these genes were detected (Figures 1 and 3). PITX2 was examined for potential rearrangements based on previous case reports of its association with patients with hypoplasia or agenesis of the corpus callosum.4,25 The PITX2 gene is estimated to map about 1.7 Mb proximal to the translocation breakpoint junction between the markers D4S2449 and Q9H1 at 4q25. Using a panel of amplimers mapping to specific exons of the PITX2 gene, a deletion was found from exons 6–7 on flow-sorted derivative 4 and 14 chromosomes of this patient, an estimated interval of at least 3.9 kb. Exons 1–5 were still present on the derivative 4 chromosome (Figure 4a and b). Densitometric analysis was used to confirm these findings within the PITX2 gene. Specifically, the 5′ end of the PITX2 gene was present in two copies and the 3′ end was present in one copy (Figure 4a and c). The marker R380DK4, mapping about 25 kb telomeric to PITX2 was found to be deleted, and the marker R380DK3 mapping about 50 kb telomeric to PITX2 not deleted, suggesting a microdeletion as large as 50 kb commencing within the 3′ end of PITX2 and extending towards the telomere.
Analyses of the PITX2 gene for rearrangements. (a) Schematic of the PITX2 gene with map positions of markers and probes (b) Flow-sorted chromosome analysis with amplimers mapping within the PITX2 gene. (c) Densitometry analysis with probes mapping to the 5′ or 3′ regions of PITX2. The intensity values are in counts per minute.
Discussion
The translocation breakpoint junctions of this patient were defined on chromosomes 4q25 and 14q13, by mapping on flow-sorted aberrant chromosomes. Analyses of candidate genes within a 2 Mb interval at the 14q13 and 4q25 breakpoint junctions showed no deletions or rearrangements in PRO0916, T2BP, ANK2, SLC25A21, NKX2.1 and NKX2.9 thus excluding them as potentially contributing to the phenotype in this patient. The possibility of long-range position effects exerted on the transcriptional regulation of these genes, however, cannot be excluded. The translocation breakpoint junction of this patient was found within the third intron of the MIPOL1 gene at 14q13, and on chromosome 4q25 between the markers D4S2449 and Q9H1. In conjunction with this finding, a microdeletion as large as 50 kb was found within and distal to the 3′ end of the PITX2 gene. Specifically, as part of the microdeletion on 4q25, at least a 3.9 kb deletion that deleted exons 6 and 7 of the PITX2 gene was found. The PITX2 gene mapped about 1.7 Mb centromeric to D4S2449 and Q9H1. A translocation breakpoint in conjunction with a neighboring deletion implied that the interval at 4q25 was possibly inverted in a complex microrearrangement. In this manner, there are two possible models of a complex microrearrangement. In the first model, an unbalanced translocation breakpoint within the PITX2 gene fused with the MIOPL1 gene, followed by an inversion of the interval between the 3′ end of the PITX2 gene and the markers D4S2449 and Q9H1. In the second model, a translocation breakpoint between the markers D4S2449 and Q9H1 and a fusion with the MIOPL1 gene were followed by an inversion of a proximal 1.7 Mb interval that disrupted and deleted part of the 3′ end of the PITX2 gene. This rearrangement was unlikely to be detected in the proband's karyotype because of insufficient metaphase GTG banding resolution. Moreover, a 1.7 Mb region is too small an interval to contain multiple distinguishable cytogenetic bands required to ascertain an inversion.
Most remarkable is the finding of a patient affected with tetramelic mirror image polydactyly and no central nervous system anomalies, and a disruption of the 11th intron of the MIPOL1 gene.6 Since the C-terminal region of the MIPOL1 protein has thus far been postulated to contain functional domains such as coiled-coil domains,6 it is likely that the translocation breakpoint junction of the tetramelic mirror image polydactyly patient affected the coding potential and function of the disrupted protein. Our patient who had no tetramelic mirror image polydactyly was demonstrated to contain a translocation breakpoint junction in the 5′ UTR of the MIPOL1 gene. Given this finding, the disrupted MIPOL1 gene in our patient is likely to be functional since the coding potential of the protein is not affected. A promoter within the third intron of the disrupted MIPOL1 gene at 14q13 or even a promoter on 4q25 can serve to regulate normal transcription of this disrupted gene thereby explaining why our patient did not have any phenotypes in common with the other patient.6 Two other reported patients1 also had translocation breakpoints at 14q13 that mapped within 300 kb of our patient. These patients, however, had no features in common with each other or with our patient. Furthermore, the disease genes for these other cases are unidentified.
We hypothesize that the deletion of exons 6 and 7 of the PITX2 gene is more likely the cause of the phenotypic spectrum in our patient. In our patient, the 3′ ends of all four PITX2 transcript isoforms were affected, resulting in deletion of the entire C-terminal domain and the homeodomain in the encoded proteins. The three promoters remained intact. No other genes are found within the estimated 50 kb microdeletion at the 3′ end of the PITX2 gene. The microrearrangement found within the PITX2 gene is predicted to produce a null allele. Hence, haploinsufficiency of the PITX2 gene is likely the cause of the phenotype. Functional studies performed on a selection of Axenfeld–Rieger syndrome type 1 disease causing mutations show these mutations reduced slightly or completely abrogate the transactivation or binding ability of the PITX2 protein.26,27 Furthermore, one PITX2 homeodomain mutation caused slight cytoplasmic mislocalization of the protein.31 Based on these findings, it was proposed that Axenfeld–Rieger syndrome type 1 patients may have only one copy or hyperactive copy in addition to a functional PITX2 to cause an abnormal phenotype. None of the panel of ARS1 patients tested for PITX2 mutations had MRI or CT cranial imaging reported. Thus, it is unknown whether these patients had acallosal defects as observed in our patient. Our patient did not exhibit any dental anomalies at the age of 19 months; however, it is unknown whether he would have developed glaucoma since he died during his early childhood. Many other patients with PITX2 mutations may also be nonpenetrant for the ocular and dental anomalies that are mainly associated with the Axenfeld–Rieger syndrome type 1 phenotypic spectrum, and obviously would not be initially selected for PITX2 mutation screening.
Patients with 4q25 translocations and deletions have provided evidence of the phenotypic spectrum which includes those seen in our patient. The first reported case that possibly associates PITX2 with acallosal defects was a t(1;4)(q23.1;q25) patient with Axenfeld–Rieger ocular and umbilical defects.4 This patient also had central nervous system midline defects that included hypoplasia of the corpus callosum and agenesis of the anterior commissure. Other translocation cases that mapped within 100–200 kb distal to the PITX2 gene are affected with the Axenfeld–Reiger syndrome type 1 spectrum.2,3 Among these cases, the PITX2 gene was not reported rearranged or deleted. The translocation breakpoint junctions in these cases may have exerted position effects on the regulation of the PITX2 gene. Extreme clinical heterogeneity is observed in the 4q25 deletion cases, that include Axenfeld–Rieger syndrome type 1 features, acallosal defects and craniofacial anomalies.25,28 Of the total of 12 cases reported, only five had typical Axenfeld–Rieger syndrome type 1 features. Three cases had complete deletions of the 4q25 band with no Axenfeld–Rieger syndrome type 1 phenotypes but a spectrum of CNS anomalies.25 The majority of deletion cases involving the 4q25 band have clinical features in common with our patient, who has only one functional copy of PITX2.
The PITX2 gene is proposed to function downstream of the sonic hedgehog and nodal pathway in the left–right asymmetry pathway.29 Null alleles of the Pitx2 gene were created in mice by several groups30,31,32. Heterozygous and homozygous null Pitx2 mice showed defects in the morphogenesis of the heart, mandibular and maxillary facial prominences, tooth, eye and pituitary; and in establishment of asymmetry. In fact, many features of the Pitx2−/− mice were common with those of Axenfeld–Rieger syndrome type 1 patients. However, no acallosal defects were seen among these mice since the concepti died before E15. The corpus callosum forms between E16 and E18 in the mice. A hypomorphic allele of the Pitx2 gene was also created with homozygous and heterozygous mice that were born to term.33 These mice again had features similar to the null Pitx2 mice and Axenfeld–Rieger syndrome type 1 patients. However, no reports of acallosal defects were noted. It is possible that the difference in phenotype observed between mouse and human is a result of different roles of PITX2 in these two species, or additional compensatory genes. The murine Pitx2 gene is expressed in many tissues including the brain, heart, pituitary, mandibular and maxillary regions, eye and umbilicus.34 It is expressed in the forebrain and in the midline ventral domains of the early developing forebrain.34 The murine expression pattern of Pitx2 suggests possible influences upon the midline-laterality determination by feedback control mechanisms with other midline genes such as sonic hedgehog, no tail and floating head.
In summary, we have characterized the translocation breakpoint junctions on chromosomes 4q25 and 14q13 in a patient affected with central nervous system acallosal defects. A deletion was found within the 3′ end of the PITX2 gene in conjunction with a distal breakpoint junction about 1.7 Mb away, suggesting a microinversion between the PITX2 gene and the translocation breakpoint junction on chromosome 4q25. A breakpoint junction within the 5′ UTR of the MIPOL1 gene at 14q13 was also found. Our findings are of importance in understanding the role of the PITX2 gene in human development, particularly, the link between the midline and laterality developmental pathways in the genesis of a phenotype. MRI or CT cranial imaging of patients with Axenfeld–Rieger syndrome type 1 could help in understanding the role of the PITX2 gene in human brain development.
References
Kamnasaran D, O'Brien PC, Schuffenhauer S et al: Defining the breakpoints of proximal chromosome 14q rearrangements in nine patients using flow-sorted chromosomes. Am J Med Genet 2001; 102: 173–182.
Datson NA, Semina E, van Staalduinen AA et al: Closing in on the Rieger syndrome gene on 4q25: mapping translocation breakpoints within a 50-kb region. Am J Hum Genet 1996; 59: 1297–1305.
Flomen RH, Vatcheva R, Gorman PA et al: Construction and analysis of a sequence-ready map in 4q25: Rieger syndrome can be caused by haploinsufficiency of RIEG, but also by chromosome breaks approximately 90 kb upstream of this gene. Genomics 1998; 47: 409–413.
Makita Y, Masuno M, Imaizumi K et al: Rieger syndrome with de novo reciprocal translocation t(1;4) (q23.1;q25). Am J Med Genet 1995; 57: 19–21.
Wang Q, Timur AA, Szafranski P et al: Identification and molecular characterization of de novo translocation t(8;14)(q22.3;q13) associated with a vascular and tissue overgrowth syndrome. Cytogenet Cell Genet 2001; 95: 183–188.
Kondoh S, Sugawara H, Harada N et al: A novel gene is disrupted at a 14q13 breakpoint of t(2;14) in a patient with mirror-image polydactyly of hands and feet. J Hum Genet 2002; 47: 136–139.
Kamnasaran D, Cox DW : Current status of human chromosome 14. J Med Genet 2002; 39: 81–90.
Cox CJ, Espinoza HM, McWilliams B et al: Differential regulation of gene expression by PITX2 isoforms. J Biol Chem 2002; 277: 25001–25010.
Yu X, St Amand TR, Wang S et al: Differential expression and functional analysis of Pitx2 isoforms in regulation of heart looping in the chick. Development 2001; 128: 1005–1013.
Schweickert A, Campione1 M, Steinbeisser H, Blum M : Pitx2 isoforms: involvement of Pitx2c but not Pitx2a or Pitx2b in vertebrate left–right asymmetry. Mech Dev 2000; 90: 41–51.
Mucchielli ML, Mitsiadis TA, Raffo S, Brunet JF, Proust JP, Goridis C : Mouse Otlx2/RIEG expression in the odontogenic epithelium precedes tooth initiation and requires mesenchyme-derived signals for its maintenance. Dev Biol 1997; 189: 275–284.
Degar BA, Baskaran N, Hulspas R, Quesenberry PJ, Weissman SM, Forget BG : The homeodomain gene Pitx2 is expressed in primitive hematopoietic stem/progenitor cells but not in their differentiated progeny. Exp Hematol 2001; 29: 894–902.
Arakawa H, Nakamura T, Zhadanov AB et al: Identification and characterization of the ARP1 gene, a target for the human acute leukemia ALL1 gene. Proc Natl Acad Sci USA 1998; 95: 4573–4578.
Wei Q, Adelstein RS : Pitx2a expression alters actin–myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell 2002; 13: 683–697.
Semina EV, Reiter R, Leysens NJ et al: Cloning and characterization of a novel bicoid-related homeobox trans-cription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 1996; 14: 392–399.
Doward W, Perveen R, Lloyd IC, Ridgway AE, Wilson L, Black GC : A mutation in the RIEG1 gene associated with Peters' anomaly. J Med Genet 1999; 36: 152–155.
Kulak SC, Kozlowski K, Semina EV, Pearce WG, Walter MA : Mutation in the RIEG1 gene in patients with iridogo-niodysgenesis syndrome. Hum Mol Genet 1998; 7: 1113–1117.
Telenius H, Pelmear A, Tunnacliffe A et al: Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 1992; 4: 257–263.
Matsumoto N, Soeda E, Ohashi H et al: A 1.2 megabase BAC/PAC contig spanning the 14q13 breakpoint of t(2;14) in a mirror-image polydactyly patient. Genomics 1997; 45: 11–16.
Fiermonte G, Dolce V, Palmieri L et al: Identification of the human mitochondrial oxodicarboxylate carrier. Bacterial expression reconstitution, functional characterization, tissue distribution and chromosomal location. J Biol Chem 2001; 276: 8225–8230.
Kimura S, Hara Y, Pineau T et al: The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev 1995; 10: 60–69.
Pabst O, Herbrand H, Arnold HH : Nkx2-9 is a novel homeobox transcription factor which demarcates ventral domains in the developing mouse CNS. Mech Dev 1998; 73: 85–93.
Bouley M, Tian MZ, Paisley K, Shen YC, Malhotra JD, Hortsch M : The L1-type cell adhesion molecule neuroglian influences the stability of neural ankyrin in the Drosophila embryo but not its axonal localization. J Neurosci 2000; 20: 4515–4523.
Chan W, Kordeli E, Bennett V : 440-kD ankyrinB: structure of the major developmentally regulated domain and selective localization in unmyelinated axons. J Cell Biol 1993; 123: 1463–1473.
Kulharya AS, Maberry M, Kukolich MK et al: Interstitial deletions 4q21.1q25 and 4q25q27: phenotypic variability and relation to Rieger anomaly. Am J Med Genet 1995; 55: 165–170.
Kozlowski K, Walter MA : Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Hum Mol Genet 2000; 9: 2131–2139.
Priston M, Kozlowski K, Gill D et al: Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld–Rieger syndrome. Hum Mol Genet 2001; 10: 1631–1638.
Schinzel A, Brecevic L, Dutly F, Baumer A, Binkert F, Largo RH : Multiple congenital anomalies including the Rieger eye malformation in a boy with interstitial deletion of (4) (q25-->q27) secondary to a balanced insertion in his normal father: evidence for haplotype insufficiency causing the Rieger malformation. J Med Genet 1997; 34: 1012–1014.
Roessler E, Muenke M : Midline and laterality defects: left and right meet in the middle. Bioessays 2001; 23: 888–900.
Lin CR, Kioussi C, O'Connell S et al: Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature 1999; 401: 279–282.
Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF : Function of Rieger syndrome gene in left right asymmetry and craniofacial development. Nature 1999; 401: 276–278.
Kitamura K, Miura H, Miyagawa-Tomita S et al: Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 1999; 126: 5749–5758.
Gage PJ, Suh H, Camper SA : Dosage requirement of Pitx2 for development of multiple organs. Development 1999; 126: 4643–4651.
Muccielli ML, Martinez S, Pattyn A, Goridis C, Brunet JF : Otlx2, an Otx-related homeobox gene expressed in the pituitary gland and in a restricted pattern in the forebrain. Mol Cell Neurosci 1996; 8: 258–271.
Acknowledgements
We thank Dr Michael Walter for discussions of PITX2. This study is funded by Grant 6-FYO1-267 of March of Dimes Birth Defects Foundation to DWC. DK holds scholarships from Alberta Heritage Foundation for Medical Research and Canadian Institutes of Health Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kamnasaran, D., O' Brien, P., Zackai, E. et al. Rearrangement in the PITX2 and MIPOL1 genes in a patient with a t(4;14) chromosome. Eur J Hum Genet 11, 315–324 (2003). https://doi.org/10.1038/sj.ejhg.5200963
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200963
Keywords
This article is cited by
-
CD105 (Endoglin) exerts prognostic effects via its role in the microvascular niche of paediatric high grade glioma
Acta Neuropathologica (2012)
-
Axenfeld–Rieger syndrome and spectrum of PITX2 and FOXC1 mutations
European Journal of Human Genetics (2009)