Abstract
The Williams-Beuren syndrome (WBS) is a complex developmental disorder with multisystemic manifestations including supravalvular aortic stenosis (SVAS), a so-called elfin face, a hoarse voice, and a specific cognitive phenotype. Most WBS patients have a >1 Mb deletion on one of their chromosomes 7 in q11 but except for elastin, whose haploinsufficiency causes the cardiovascular malformations, it is unknown which genes in the deletion area contribute to the phenotype. We have investigated a family with a cytogenetically balanced translocation t(7;16)(q11.23;q13) in which affected individuals manifested a broad spectrum of clinical phenotypes ranging from a hoarse voice as the only feature to the full WBS phenotype. Molecular cytogenetic and DNA sequence analyses of the translocation breakpoint showed that the cytogenetic rearrangement disrupts the elastin gene locus within intron 5 in the exact same manner in all translocation carriers. The recently described large inversion of the 7q11.23 region was not present in this family. Our data demonstrate that disruption of the elastin gene by a translocation breakpoint may cause classical WBS, atypical WBS, SVAS, or no recognisable phenotype, and provide a clear example for extensive phenotypic variability associated with a position effect in humans.
Similar content being viewed by others
Introduction
Williams-Beuren syndrome (WBS) (OMIM 194050; http://www.ncbi.nlm.nih.gov:80/entrez/dispomim.cgi?id=194050) is a rare (1/20000–1/50000), generally sporadic and complex developmental disorder with multisystemic manifestations, which in most patients is caused by the hemizygous deletion of a large genome region on chromosome 7q11.23. The commonly observed deletions are quite homogeneous in size. The most frequently deleted segment (≈1.5–1.7 Mb) extends between two highly homologous duplicons of 320–500 kb, containing the polymorphic markers D7S489B and D7S489A and harbours at least 19 genes including the elastin (ELN) locus.1,2,3,4 These duplicons include about 30 kb of the 3′-region of a transcribed gene, called GTF2I (telomeric copy) and GTF2IPI (centromeric copy). The high conservation (99.9%) of the coding sequences of GTF2I and GTF2IPI, as well as the preservation of restriction fragment patterns at both loci, indicate that the duplication is of recent evolutionary origin.5 With probes containing parts of the elastin gene the deletion can be readily detected by fluorescence in situ hybridisation (FISH) in 90–99% of individuals with WBS.6,7,8,9,10,11 It has been postulated that a number of 7q11.23 deletions occur in association with an interchromosomal rearrangement, indicative of an unequal crossing-over event between mispaired repeats of the two homologous chromosomes 7.12,13 Clinical symptoms of the WBS phenotype include growth retardation, congenital cardiovascular disease (supravalvular aortic stenosis (SVAS) and peripheral pulmonal stenosis (PPS)), characteristic dysmorphic facial features (‘elfin face’), frequent dental abnormalities, a hoarse voice, mental retardation or learning difficulties, a unique cognitive profile and a distinct personality.2,14,15,16,17, With the exception of the cardiovascular aspects of the disorder and possibly the hoarse voice, which are supposed to be caused by haploinsufficiency for elastin (ELN), the role of the genes contributing to the other features of WBS is still undetermined. No other of the WBS features is seen in patients with isolated ELN deficiency.18,19 The identification of individuals with smaller deletions and other aberrations may contribute to elucidate the pathogenesis of WBS and help to understand which of the genes in the common WBS deletion are dosage sensitive and contribute to particular features of the phenotype.3
Here we report a family with a cytogenetically balanced translocation t(7;16)(q11.23;q13). Translocation carriers from the family, which was first described in 1979,20 presented with an extremely variable phenotype ranging from full WBS to minimal signs which were only diagnostic in the familial context. Cloning of the translocation breakpoint revealed that the cytogenetic rearrangement disrupts the elastin gene locus in all translocation carriers. The five affected members of our family present a group of individuals with the same disruption of ELN but a wide variation in the phenotype, which is likely to reflect a position variegation effect.
Subjects and methods
Eight members of a family with the clinical suspicion of familial WBS20 were reinvestigated by clinical, cytogenetic, molecular cytogenetic, and molecular genetic methods (Figure 1). Clinical investigation included an extensive medical history, physical examination, ultrasonography of the heart and kidneys, and a clinical laboratory analysis of blood. The probands were inspected for clinical features of WBS according to published criteria for clinical diagnosis of WBS17 and for the clinical features listed in OMIM (OMIM 194050; http://www.ncbi.nlm.nih.gov:80/entrez/dispomim.cgi?id=194050). One female was investigated only cytogenetically. For all individuals participating in this study, informed consent was obtained.
Pedigree of Williams-Beuren syndrome family with t(7;16). Individuals with t(7;16) and features of Williams Beuren syndrome (solid square and circle); Individuals with normal 46,XY or 46,XY karyotype (hatched box and circle); Individuals not investigated (open box and circle); 5 further siblings not available for investigation (open diamond).
Chromosome analysis
Peripheral blood samples were obtained from nine members of the family for chromosome analysis. For high-resolution chromosomal analysis, whole blood lymphocyte cultures were stimulated by Phythaemagglutinin (PHA) and synchronised by Methotrexate by means of standard protocols. Metaphase chromosomes were routinely analysed by standard Trypsin–Giemsa (GTG) and Quinacrine (QFQ) banding.
Fluorescence in situ Hybridisation (FISH)
FISH was performed with the LSI® Williams Syndrome Region DNA FISH Probe (VYSIS) according to the manufacturer's protocols and with the BAC CTB-51J22 (GenBank AC005056) from the commonly deleted WBS region. Additionally we used BAC CTA-208H19 and PAC RO5-1186P10 from within the common WBS-deletion interval and the flanking BACs RP11-815K3 (centromeric) and CTB-139P11 (telomeric) to detect possible inversion of the WBS region.21 The clones were biotin- or digoxigenin-labelled by nick translation. Chromosomal in situ suppression hybridisation and detection of the biotinylated probe was carried out as described.22 In each translocation carrier at least 20 metaphases and 100 interphases were examined.
Construction of human-hamster somatic cell hybrids
The HGPRT negative Chinese hamster cell line RJK88 (Coriell Cell Repositories, Camden, USA, No. NA10658), was fused with Polyethlenglycol (PEG) in equal proportion with human lymphoblastoid cells from the affected individual III/7 as described earlier.23 Individual hybrid clones were analysed by PCR using the chromosome 7 markers D7S544E and D7S557E for the presence of chromosome 7 or its derivatives. Clones containing either the marker from the telomeric end of 7q or from 7p, respectively, were selected for further analysis. Presence of a single derivative chromosome was verified by FISH analysis on metaphase chromosomes, by means of chromosome 7 and chromosome 16-specific paint probes (Cambio).
Southern analysis
Restriction enzyme digestion of genomic DNA was carried out as recommended by the manufacturer (MBI Fermentas) with the exception that a 10-fold excess of enzyme be used. The digested DNA (10 μg) was separated on 0.8% agarose gels in 1× TBE buffer. Gels were depurinated for 20 min in 0.25 M HCl, rinsed briefly in H2O, soaked in denaturating buffer (0.5 M NaOH, 1.5 M NaCl) for 30 min and incubated for 30 min in neutralisation buffer (0.5 M Tris HCl, pH 7.0). Capillary transfer to nylon membranes (Pall Biodyne A 0.2 μm) was carried out overnight in 10×SSC. The membrane was allowed to dry briefly and then UV-cross linked (UV Stratalinker 2400, Stratagene).
For hybridisation, a radioactive DNA probe was prepared to high specific activity, >2×109 c.p.m. per microgram of DNA, by the Rediprime II labelling system (Amersham Pharmacia) according to the manufacturer's instructions. Membranes were prehybridised in 5 ml/cm2 Rapidhyb buffer Solution (Amersham Pharmacia) at 65°C for 30 min. The radioactive probes were added in the same solution to a >1×106 cpm/ml. All hybridisations were carried out using standard procedures. Briefly, hybridisation at 65°C for 1–2.5 h, two washes in 2×SSC 0.1%, SDS for 10 min at 25°C were followed by two stringency washes in 0.1 SSC 0.1% SDS at 65°C for 10 min at 65°C. Filters were exposed to X-ray film (Kodak Biomax MR) overnight at −80°C with two intensifying screens.
Cloning of the translocation breakpoint
Ten μg of patient's genomic DNA were digested with SacI and size selected on a 0.8% agarose gel. The 1.0 and 1.2 kb fractions containing the normal and the rearranged fragment, respectively, were purified (QIAEX II kit) and ligated into SacI digested and dephosphorylated.
pBluescript II SK(+) (Stratagene). Specific ligation products were amplified using the vector specific primer M13F(−21) and a chromosome 7 insert specific primer A. The PCR products were used in 1 : 1250 dilution for a second round of PCR using a chromosome 7 specific nested primer B and M13F(−21). The resulting 1.2 and 1.0 kb products were sequenced directly after gel purification.
PCR amplification
DNA clone inserts, somatic cell hybrid DNA and total human DNA samples were amplified by PCR as described.24 Genomic DNA (100 ng) or plasmid DNA (10 ng) was amplified in a 25 μl reaction containing 20 pmol of each oligonucleotide primer and ready-to-go beads (Amersham Pharmacia).
Amplification conditions were 94°C for 3 min followed by 35 cycles of 60°C for 30 s, 72°C for 60 s and 94°C of 30 s (primers are listed in Table 2).
Sequencing and sequence analysis
Sequencing reactions were performed on an ABI 310 sequencer using an ABI PRISM Dye terminator cycle sequencing ready reaction kit (Perkin Elmer) and vector specific or sequence derived primers. Sequence analysis was performed with the BLAST program at the NCBI using the databases NR and HTGS.25
Results
Clinical analysis
Clinical investigation of the five translocation carriers (two males, three females) revealed a wide variation in their phenotypes (Table 1). One female patient (III/7, Figure 1) with clinical unambiguous WBS presented with typical dysmorphic facies (broad forehead, full cheeks, epicanthus, medial eyebrow flare, depressed nasal bridge, anteverted nares, broad nasal tip, open mouth, protruding lower lip) (Figure 2a,b), ocular anomalies, distinct dental abnormalities (Figure 2c), short stature (5th–10th percentile), mild scoliosis, valvular aortic stenosis, peripheral pulmonal stenosis, WBS personality with overfriendliness, attention deficit disorder anxiety and hypersensitivity to sound, and mild mental retardation (IQ 66) (see Table 1). The only identifiable feature associated with WBS in her mother (II/3) was a hoarse voice (Figure 2d). Hence this patient is essentially asymptomatic and would not have been identified as a translocation carrier without the familial context. Patient III/1 presented with severe SVAS, that needed surgical treatment, PPS and renal cysts of the left kidney but neither facial signs nor cognitive impairment or a behavioural phenotype. His brother, (III/4) showed mild SVAS and trivial PPS, a malocclusion of teeth and blue iris. The mother of the two boys (II/2) showed mild facial features of WBS, blue iris, and a malocclusion. Educational history of the mentally non-retarded translocation carriers (II/2 and II/3 and III/1 and III/4) revealed normal schooling. The two brothers (III/1 and III/4) finished high school and work as accountant and bank employee, respectively. Psychological testing regarding the characteristic WBS cognitive profile14,16 was not possible in these individuals due to unavailability of this test in the German language.
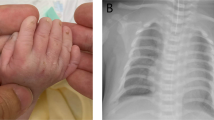
Facial appearance of the proposita (III/7) with t(7;16) and her mother (II/3) with t(7;16). Portrait of the proposita (III/7) at the age of 8 years (A; reprinted by permission20) and 29 years (B), showing the elfin face with broad forehead, full cheeks, epicanthus, medial eyebrow flare, depressed nasal bridge, anteverted nares, broad nasal tip, open mouth and protruding lower lip. The dental anomalies in the proposita (III/7) at the age of 8 years are shown in (C; reprinted by permission20). Note the absence of facial dysmorphism in the mother of the proposita (D), who has a hoarse voice as the only WBS feature.
Chromosome analysis
Chromosome analysis revealed a cytogenetically balanced translocation, t(7;16) (q11.23;q13) in five individuals and a normal karyotype in four individuals (see Figures 1 and 3).
Fluorescence in situ hybridisation analysis (FISH)
FISH with the LSI® Williams Syndrome Region DNA FISH probe (VYSIS) on inter- and metaphases from all five translocation carriers revealed two red and two green signals without split of the ELN specific signal. The distribution of the signals to the translocation chromosomes showed a green signal on the normal chromosome 7 and the derivative chromosome 16 and a red signal on the normal chromosome 7 and the derivative chromosome 16 suggesting the breakpoint lying proximal from ELN. The probands with normal karyotype served as controls. Hybridisation with BAC CTB-51J22 (GenBank AC005056), which extends more proximal over ELN than the LSI® Williams Syndrome Region DNA FISH probe (VYSIS) revealed a split hybridisation signal on the derivative chromosomes 7 and 16 indicating that the breakpoint could reside within ELN (data not shown).
We also tested by FISH whether an inversion of the WBS region recently described in atypical WBS patients with chromosomal rearrangements and some parents of WBS patients with the 1.5 Mb deletion is present in translocation carriers of our family. Two different triple combinations of the probes 815k3, 208H19, 1186P10, 139P11 were used for three-colour FISH analysis of the WBS critical region.21 On metaphase chromosomes the t(7;16) breakpoint split these probe sets into the two expected colour combinations to the derivative chromosomes indicating that no inversion is present (Figure 4a,d). On interphase nuclei signals of both probe mixtures also appeared in the expected order on the normal chromosome 7 (Figure 4b,c,e,f). Thus, the combined FISH results indicate that no predisposing inversion of the WBS region has occurred prior to the translocation in our family.
Exclusion of a WBS inversion by three-colour FISH on metaphase chromosomes of the index patient carrying a t(7;16)(q11.23;q12.1). The localisation of probes (coloured dots), elastin gene (ELN), and translocation breakpoint (arrow), and the colour-label and probe order from centromer to telomer for the two probe sets (a,b) is given (the white boxes along the line represent duplicons mediating the described 1.5 Mb inversion). With the probe set shown in (a) we observed a single green signal on the derivative chromosome 7 whereas the inversion is expected to create a mixed (yellow) signal on the der(7), and the probe set shown in (b) revealed mixed signals on both derivative chromosomes whereas the inversion is expected to create a single green signal on the derivative chromosome 16 (note that the yellow signal appears as a mixed red-green signal).
Physical mapping of the translocation breakpoint
The two derivative translocation chromosomes der(7) and der(16) from patient III/7 were separated in human/Chinese hamster somatic cell hybrids from each other and the intact chromosome 7. Segregation analysis of STS markers D7S2233, Eln1, Eln2, Eln3, Eln4, Eln5, Eln6, Eln7, Eln8, Eln9, Eln10, Eln11, LimKE1 and D7S2472 (Table 2) mapping within the common WBS deletion in hybrid clones RJK-PM7 and RJK-PM18, retaining the der(7) or the der(16) chromosome only, indicated that the translocation had broken chromosome 7 between exon 4 and 5 of the elastin gene as depicted in Table 3.
Cloning of the translocation breakpoint
Southern blot analysis of DNA from patient III-7 digested with SacI and hybridised with a probe flanking the breakpoint on chromosome 7 within intron 5 showed a 1.2 kb fragment in addition to the usual 1.0 kb fragment (Figure 5). The additional putative junction fragment was cloned, PCR amplified and sequenced. BLAST analysis of the DNA sequence against the NR and HTGS databases at the NCBI identified sequence identities with two clones CTB-51J22 and RP11-405F3, which were mapped to 7q11.23 and 16q13, respectively. DNA sequence analysis showed that the breakpoint on chromosome 7 was indeed within intron 5 of the elastin gene as predicted from the cell hybrid analysis and on chromosome 16 within intron 1 of the TM7XN1 gene (Figure 6).
(A) Sequence analysis of the t(7;16)(q11.23;q13) breakpoints. The sequences of chromosome 7 and 16 were obtained from GenBank (AC005056; AC018552). Cloning and sequencing of an aberrant restriction fragment obtained the sequence of the der(16). The sequence of the der(7) was obtained by direct sequencing of PCR products spanning the breakpoint. In the course of the rearrangement no base pair was lost from either the chromosome 7 or chromosome 16 sequences. (B) Genomic structure of the breakpoint region in 7q11.23. Oligonucleotide primers used for PCR amplification of fragments flanking or spanning the breakpoint and the location of the SacI restriction site are underlined and in boldface. The basepairs corresponding to exon 5 of the elastin gene are boxed. A dotted line indicates the positions of the SacI restriction sites. The position of the breakpoint is underlined.
Identification of a translocation specific junction fragment (A) Schematic overview of the genomic structure of elastin in the breakpoint region. Exons (E) are represented by boldface rectangles. The boldface bar placed above the corresponding genomic structure of the gene represents the genomic probe used for the Southern analysis shown in (B). (A, B, C, D) representing primers used to amplify fragments covering the breakpoint. (B) Southern blot hybridisation of the genomic DNA probe against genomic DNA of a control patient GM3104 (lane 2) and the translocation patient PM (lane 1) digested with SacI. An additional restriction fragment (arrow) can be observed in lane 1 representing the junction fragment from the der(16). For details of the chromosome 16 breakpoint see Figure 5A
Analysis of the translocation breakpoint in the other carriers
To determine if the rearrangement is identical in all carriers or whether differences in the breakpoints are responsible for the heterogeneous phenotype of the affected family members, fragments covering the breakpoints were amplified by PCR using primers B, C, 16for and 16rev, respectively (Table 2) and sequenced. All five translocation carriers showed PCR fragments of the same size (data not shown). Direct sequencing of the fragments indicated that the chromosomal breakpoints in all five translocation carriers were identical.
Discussion
WBS is generally sporadic and usually caused by the hemizygous deletion of about 1.5–1.7 Mb on chromosome 7q11.23, which harbours at least 19 genes including the elastin gene. Although a number of genes within the commonly deleted WBS region are discussed to contribute to particular features of the WBS phenotype, the main question, which gene is responsible for which part of the phenotype remains unanswered except for elastin. One approach to solve the problem is the identification and molecular characterisation of individuals with small deletions within the common WBS deletion. These studies have however not resulted in an unequivocal gene-phenotype correlation.18 Another is the study of carriers of balanced translocations and a WBS-like phenotype.
Balanced translocations are rare causes of monogenic disorders but have guided the cloning of the disease-causing gene in several instances.26,27,28 Most of these translocations are sporadic and disrupt the disease gene or neighbouring sequences. The present study describes the mapping and DNA sequencing of the breakpoints of a cytogenetically balanced familial translocation t(7;16)(q11.23;q13) associated with WBS. There are two reports of chromosomal translocations involving the 7q11.23 region associated with SVAS and ‘atypical’ WBS.29 One sporadic patient with a translocation t(6;7)(q27;q11.23), SVAS and severe hydrops fetalis has been reported.29 Based on FISH-analysis the chromosome 7 breakpoint in this patient was speculated to disrupt the elastin gene but no rigorous molecular evidence was provided. Further it was not possible to distinguish whether this patient had isolated SVAS or WBS given the degree of body oedema and the early gestational age of the patient. Curran et al.30 and Morris et al.31 reported a family with dominant inheritance of SVAS and a familial translocation t(6;7)(p21.1;q11.23). They demonstrated that the elastin gene was disrupted in exon 28 by the translocation. All affected family members had isolated SVAS with one exception. One translocation carrier was reported to have full cheeks, a hoarse voice, and fifth finger clinodactyly and may be classified as a partial or ‘atypical’ WBS patient, much like some of the translocation carriers in the family described here (Table 1). However the proposita in our family (III/7) had classical WBS (Figure 2, Table 1) whereas her mother (II/3) who carries the same translocation is asymptomatic. The only WBS-associated symptom was a hoarse voice, which is only diagnostic in the familial context. Hence the phenotypic variability in our family is larger than in the family reported by Curran et al.30
DNA sequence analyses of the translocation breakpoint in the family described here showed that the cytogenetic rearrangements disrupted the elastin gene locus within intron 5 and the TM7XN1 gene locus on chromosome 16 within intron 1 in exactly the same manner in all translocation carriers. Mutations in ELN result in isolated SVAS, one of the clinical features of WBS. Haploinsufficiency of elastin caused by the 1.5 Mb WBS deletion is responsible for the SVAS in WBS but can not explain the other features of WBS, in particular the cognitive phenotype and dysmorphic facial appearance associated with the WBS deletions but not with elastin mutations. Therefore the expected phenotype in our family is SVAS, not WBS. Why then do the translocation carriers in our family develop features of WBS beyond SVAS and why is the phenotype in carriers with essentially the same molecularly defined translocation so variable?
A position effect may cause WBS and explain the variable phenotype
The most likely explanation is that the translocation event causes a long-range position effect.32,33 There are several examples for monogenic developmental disorders, which are caused by translocations, which are believed to exert a position effect. Holoprosencephaly (HPE3), has been shown to be caused by mutations affecting the sonic hedgehog (SHH) gene34 or by translocations up to 250 kb 5′ to the gene.35 Other examples of putative positional effects in development disorders where the breakpoints were located downstream to the gene include the GLI3 gene in Greig acrocephalopolysyndactyly syndrome,27 the SOX 9 gene in campomelic dysplasia28 and PAX6 in aniridia.36 These translocations are sporadic with few exceptions. It is therefore not possible to identify phenotypic variability associated with identical translocations. It is also notable that the site of the translocation breakpoint causing the position effect in these examples could be several hundreds of kilobases away from the affected gene. In the region between the putative gene regulatory elements and the gene itself no additional open reading frames were identified. In contrast the common WBS deletion region is gene rich with about one gene in 40 kb. Long-range effects on other genes as a consequence of the translocation within the ELN locus are thus not easily explained.
One of the most intriguing aspects of our report is the large phenotypic variability between the translocation carriers which ranges from asymptomatic (II/3), partial WBS with SVAS (III/1, III/4) or without SVAS (II/2) to the full WBS phenotype (III/7).
While this may be explained by unidentified genetic differences between family members, environmental, or stochastic factors, we favour the explanation of a position effect.
The protein coding potential of ELN/TM7XN1 transcripts
In the transcripts potentially arising from the der(16) or the der(7), the ORFs between elastin and TM7XN1 are continuous and therefore, potentially, could be translated into fusion proteins. Formal proof of the existence or non-existence of such transcripts will depend upon functional analysis. TM7XN1 encodes a protein of 693 amino acids, GPR56, that shows highest identity (32%) to HE6, a member of a subclass of the class B secretin-like G-protein-coupled receptors.37 As the physiological role of TM7XN1 is still unclear37,38 and there are no reports showing that mutations of TM7XN1 cause a distinct phenotype any influence of the ELN/TM7XN1 fusion protein on the WBS phenotype in our family is questionable. Consistent with this, no features which are not part of the typical phenotypic spectrum of WBS, were present in any of the patients. It is also not expected that a putative elastin/TM7XN1 fusion protein retains important elastin properties given the complex control and structure of elastin.39 This is supported by the presence of SVAS in members of our family.
A caveat for phenotype-genotype correlation studies
The favoured strategy to identify genes contributing to various features of the WBS phenotype is to analyse patients with small deletions in the WBS critical region and partial WBS phenotypes. Position effects of deletion breakpoints might, however, bias the results and lead to a failure of this approach.
A genomic polymorphism consisting of a large inversion of the 7q11.23 WBS region has recently been described in 27 % of the individuals with ‘atypical’ WBS without the classical 1.5 Mb deletion and in one of the parents of 33% of WBS probands carrying the 1.5 Mb deletion.21 The authors suggest that the inversion may predispose individuals to WBS-causing microdeletions or translocations. They also discuss that two of their ‘atypical’ WBS patients had the inversion as the sole detectable abnormality in the WBS region and speculate that the inversion may be causally related to the clinical phenotype in these patients.21 No 7q11.23 inversion was present in any of the family members reported here. Hence the inversion has neither predisposed to the translocation nor does it contribute to the phenotypes or phenotypic variability in our family.
In summary our family presents a group of individuals who differentially exhibit individual features of the complex multi-system WBS phenotype due to an identical balanced translocation disrupting ELN at exon 5. This suggests that other breakpoints in the WBS critical region, eg small microdeletions or inversions may also have variable effects on the phenotype making genotype-phenotype correlations difficult to establish. Our findings may have implications for strategies aimed to dissect the WBS phenotype.
References
Williams JCP, Barratt-Boyes BG, Lowe JB . Supravalvular aortic stenosis Circulation 1961 24: 1311–1318
Beuren AJ . Supravalvular aortic stenosis: a complex syndrome with and without mental retardation Birth Defects 1972 5: 45–56
Francke U . Williams-Beuren syndrome: genes and mechanisms Hum Mol Genet 1999 8: 1947–1954
Peoples R, Franke Y, Wang YK et al. A Physical Map, Including a BAC/PAC Clone Contig, of the Williams-Beuren Syndrome-Deletion Region at 7q11.23 Am J Hum Genet 2000 66: 47–68
Perez Jurado LA, Wang YK, Peoples R et al. A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK Hum Mol Genet 1998 7: 325–334
Nickerson E, Greenberg F, Keating MT, McCaskill C, Shaffer LG . Deletions of the elastin gene at 7q11.23 occur in approximately 90% of patients with Williams syndrome Am J Hum Genet 1995 56: 1156–1161
Perez Jurado LA, Peoples R, Kaplan P, Hamel BC, Francke U . Molecular definition of the chromosome 7 deletion in Williams syndrome and parent-of-origin effects on growth Am J Hum Genet 1996 59: 781–792
Brondum-Nielsen K, Beck B, Gyftodimou J et al. Investigation of deletions at 7q11.23 in 44 patients referred for Williams-Beuren syndrome, using FISH and four DNA polymorphisms Hum Genet 1997 99: 56–61
Fryssira H, Palmer R, Hallidie-Smith KA et al. Fluorescent in situ hybridisation (FISH) for hemizygous deletion at the elastin locus in patients with isolated supravalvular aortic stenosis J Med Genet 1997 34: 306–308
Hou JW, Wang JK, Wang TR . FISH analysis in both classical and atypical cases of Williams-Beuren syndrome Zhonghua Min Guo Xiao Er Ke Yi Xue Hui Za Zhi 1998 39: 398–403
Mila M, Carrio A, Sanchez A et al. Clinical characterization, molecular and FISH studies in 80 patients with clinical suspicion of Williams-Beuren syndrome Med Clin (Barc) 1999 113: 46–49
Dutly F, Schinzel A . Unequal interchromosomal rearrangements may result in elastin gene deletions causing the Williams-Beuren syndrome Hum Mol Genet 1996 5: 1893–1898
Urban Z, Helms C, Fekete G et al. 7q11.23 deletions in Williams syndrome arise as a consequence of unequal meiotic crossover Am J Hum Genet 1996 59: 958–962
Bellugi U, Bihrle A, Jernigan T, Trauner D, Doherty S . Neuropsychological, neurological, and neuroanatomical profile of Williams syndrome Am J Med Genet Suppl 1990 6: 115–125
Osborne LR . Williams-Beuren syndrome: unraveling the mysteries of a microdeletion disorder Mol Genet Metab 1999 67: 1–10
Mervis CB, Robinson BF, Bertrand J et al. The Williams syndrome cognitive profile Brain Cogn 2000 44: 604–628
Morris CA, Mervis CB . Williams Syndrome and related disorders in Lander E (ed) Annual Review of Genomics and Human Genetics Palo Alto: Annual Reviews 2000 pp. 461–484
Tassabehji M, Metcalfe K, Donnai D et al. Elastin: genomic structure and point mutations in patients with supravalvular aortic stenosis Hum Mol Genet 1997 6: 1029–1036
Metcalfe K, Rucka AK, Smoot L et al. Elastin: mutational spectrum in supravalvular aortic stenosis Eur J Hum Genet 2000 8: 955–963
Hammerer I, Gassner I, Muller W . Familiar occurrence of Elfin's face (Williams-Beurens Syndrome=wbs) and supravalvular aortic stenosis (=svas) (author's transl) Klin Padiatr 1979 191: 287–292
Osborne LR, Li M, Pober B et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome Nat Genet 2001 29: 321–325
Lichter P, Boyle AL, Cremer T, Ward DC . Analysis of genes and chromosomes by nonisotopic in situ hybridization Genet Anal Tech Appl 1991 8: 24–35
Bender K, Grzeschik KH . Possible assignment of the glyoxalase I (GLO) gene to chromosome 6 using man-mouse somatic cell hybrids Hum Genet 1976 31: 341–345
Saiki RK, Gelfand DH, Stoffel S et al. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase Science 1988 239: 487–491
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . Basic local alignment search tool J Mol Biol 1990 215: 403–410
Monaco AP, Bertelson CJ, Middlesworth W et al. Detection of deletions spanning the Duchenne muscular dystrophy locus using a tightly linked DNA segment Nature 1985 316: 842–845
Vortkamp A, Gessler M, Grzeschik KH . GLI3 zinc-finger gene interrupted by translocations in Greig syndrome families Nature 1991 352: 539–540
Wirth J, Wagner T, Meyer J et al. Translocation breakpoints in three patients with campomelic dysplasia and autosomal sex reversal map more than 130 kb from SOX9 Hum Genet 1996 97: 186–193
von Dadelszen P, Chitayat D, Winsor EJ et al. De novo 46,XX,t(6;7)(q27;q11;23) associated with severe cardiovascular manifestations characteristic of supravalvular aortic stenosis and Williams syndrome Am J Med Genet 2000 90: 270–275
Curran ME, Atkinson DL, Ewart AK et al. The elastin gene is disrupted by a translocation associated with supravalvular aortic stenosis Cell 1993 73: 159–168
Morris CA, Loker J, Ensing G, Stock AD . Supravalvular aortic stenosis cosegregates with a familial 6; 7 translocation which disrupts the elastin gene Am J Med Genet 1993 46: 737–744
Bedell MA, Jenkins NA, Copeland NG . Good genes in bad neighbourhoods Nat Genet 1996 12: 229–232
Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V . Chromatin structure and gene expression Proc Natl Acad Sci USA 1996 93: 9384–9388
Roessler E, Belloni E, Gaudenz K et al. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly Nat Genet 1996 14: 357–360
Belloni E, Muenke M, Roessler E et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly Nat Genet 1996 14: 353–356
Fantes J, Redeker B, Breen M et al. Aniridia-associated cytogenetic rearrangements suggest that a position effect may cause the mutant phenotype Hum Mol Genet 1995 4: 415–422
Liu M, Parker RM, Darby K et al. GPR56, a novel secretin-like human G-protein-coupled receptor gene Genomics 1999 55: 296–305
Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN . TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential FEBS Lett 1999 446: 292–298
Li DY, Toland AE, Boak BB et al. Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis Hum Mol Genet 1997 6: 1021–1028
Acknowledgements
We thank Drs Lucy Osborne and Steve Scherer (Department of Molecular and Medical Genetics, The University of Toronto, Ontario, Canada) for providing us with BACs CTA-208H19, RP11-815K3, CTB-139P11 and PAC R05-1186P10 for inversion screening of the WBS region. We thank Pensiri Probst, Eveline Rode and Claudia Schneider for their skilful technical assistance. This work was supported by the ‘Verein zur Förderung der Humangenetik an der Universität Innsbruck’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Duba, HC., Doll, A., Neyer, M. et al. The elastin gene is disrupted in a family with a balanced translocation t(7;16)(q11.23;q13) associated with a variable expression of the Williams-Beuren syndrome. Eur J Hum Genet 10, 351–361 (2002). https://doi.org/10.1038/sj.ejhg.5200812
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.ejhg.5200812
Keywords
This article is cited by
-
The segregation of different submicroscopic imbalances underlying the clinical variability associated with a familial karyotypically balanced translocation
Molecular Cytogenetics (2015)
-
Humangenetische Untersuchungs- und Beratungsstelle an der Landes-Frauen- und Kinderklinik Linz
Medizinische Genetik (2007)
-
Characterisation of a non-recurrent familial translocation t(7;9)(q11.23;p24.3) points to a recurrent involvement of the Williams–Beuren syndrome region in chromosomal rearrangements
Journal of Human Genetics (2006)