Abstract
The Bcl-2-family of proteins localize to intracellular membranes via a C-terminal hydrophobic membrane anchor (MA) domain, to exert their antiapoptotic or proapoptotic functions. In Drosophila, both Bcl-2 family members, DEBCL and BUFFY, contain an MA. In DEBCL the MA is necessary for the localization of protein to mitochondria and for its proapoptotic activity. BUFFY is highly similar to DEBCL but its localization and function are not clearly defined. Here, we report on comparative analysis of BUFFY and DEBCL to decipher the molecular basis for their subcellular localization. We show that these two proteins localize to distinct intracellular membranes, DEBCL predominantly to mitochondria and BUFFY to endoplasmic reticula (ER). Our results suggest that the MA-flanking residues in DEBCL, homologous to Bcl-XL, are required for the targeting of DEBCL to mitochondria. The C-terminal positively charged residues present in DEBCL are absent in BUFFY, which allows for its localization to ER. The MA in both proteins is required for the correct targeting and proapoptotic activities of these proteins. Interestingly, a functional nuclear localization signal was identified in the N-terminal region of BUFFY and in the absence of the MA, BUFFY accumulated in the nucleus. The functional implications of these findings are discussed.
Similar content being viewed by others
Main
The members of the Bcl-2 family of proteins are essential regulators of cell survival and apoptosis in metazoans.1, 2, 3, 4 The members of this Bcl-2 family share one to four Bcl-2 homology (BH) domains. The prosurvival proteins, such as Bcl-2 and Bcl–XL, are characterized by the presence of BH1-BH4 domains and act to maintain the integrity of mitochondria to prevent the release of cytochrome c.1, 2 The members of the proapoptotic Bax subfamily (Bax, Bak and Bok) contain BH1–BH3 domains and are required for mitochondrial outer membrane (MOM) permeabilization.1, 3 The BH3-only proteins such as Bid, Bad, Bik, Bim, Noxa and Puma respond to various death stimuli and induce cell death in a Bax/Bak-dependent manner or by antagonizing the prosurvival Bcl-2 proteins.1, 2
Most Bcl-2 family members contain a C-terminal hydrophobic membrane anchor (MA).1, 2, 3, 4 The MA mediates the intracellular localization of these proteins to membranes of the mitochondria, ER and nuclear envelope. MA-dependent subcellular localization is critical for the correct function of both prosurvival and proapoptotic Bcl-2 homologues.1, 2, 3, 4 In a comprehensive study, Kaufmann et al.5 described the molecular basis for the differential intracellular localization of Bcl-2 and Bcl-XL. They found that multiple positively charged residues flanking the MA of Bcl-XL that are absent in Bcl-2 appear to determine the localization of Bcl-XL to specific membrane compartments. The authors suggest that in the absence of such a strong positive charge, Bcl-2 is able to localize to other membranes as well as mitochondria.5
There are two Bcl-2 family members in Drosophila, BUFFY and DEBCL, both of which are closely related to mammalian Bok and contain BH1–BH3 domains.6, 7, 8, 9, 10, 11 Unlike in mammals, BH1–BH4 prosurvival members of Bcl-2 and the BH3-only proteins have not been reported in Drosophila. Although the precise functions of the Drosophila Bcl-2 homologues are not understood, some data suggest that BUFFY and DEBCL may have context-dependent activities.6, 7, 8, 9, 10, 11, 12 While it is well established that DEBCL largely localizes to mitochondria,7 the exact localization of BUFFY is not known. In this study, we show that BUFFY localizes primarily to endoplasmic reticula (ER). We define the residues required for appropriate localization of BUFFY and DEBCL in both mammalian and Drosophila cells. We show that while the MA is required for protein localization, the residues surrounding the MA domains of BUFFY and DEBCL significantly contribute to their localization to ER and mitochondria respectively. We also define a functional NLS in BUFFY and demonstrate that loss of the MA results in nuclear accumulation of BUFFY.
Results
BUFFY localizes to ER in an MA-dependent manner
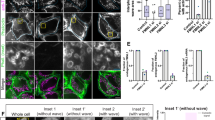
To determine the intracellular localization of BUFFY, an HA-tagged construct was transfected into COS cells. Confocal microscopy of immunostained cells demonstrated intense perinuclear staining with punctate staining in the cytoplasm, which colocalized with the ER marker, Calnexin (Figure 1a). Expression of GFP-BUFFY in Drosophila cells demonstrated a similar pattern of punctate cytoplasmic staining that colocalized with ER-Tracker but not with MitoTracker (Figure 1b). To assess the importance of the C-terminal hydrophobic MA for the correct localization of BUFFY, we expressed the protein lacking the MA (BUFFYΔMA) in Drosophila cells. In contrast to the perinuclear and punctate cytoplasmic localization of the full-length BUFFY, BUFFYΔMA failed to localize to intracellular membranes and accumulated in the nucleus (Figure 1c).
ER localization and proapoptotic activity of BUFFY are dependent on the MA. BUFFY colocalizes with the ER protein Calnexin in COS cells (a) and with ER-Tracker but not with MitoTracker in SL2 cells (b), demonstrating that BUFFY localizes to the ER. (c) Expression of transfected FLAG-BUFFY and FLAG-BUFFY ΔMA in BG2 cells. Deletion of the MA completely disrupts the localization of BUFFY, and results in protein accumulation in the nucleus. (d) BUFFY, but not a BUFFY mutant lacking the C-terminal MA (ΔMA) induces cell death when overexpressed in BG2 cells. DEBCL similarly induces apoptosis. Coexpression of both BUFFY and DEBCL enhanced cell death induced by each protein alone. Data (±S.E.M.) are from three experiments
The MA domain is essential for the proapoptotic activity of BUFFY
Overexpression of DEBCL in cells and in transgenic flies induces apoptosis.6, 7, 8, 9 In transfected cells, the proapoptotic activity of DEBCL is dependent upon its MA-dependent mitochondrial localization. To assess the ability of BUFFY to induce cell death, full-length and ΔMA mutant were expressed in Drosophila BG2 cells together with a reporter plasmid (pIE-LacZ). β-Galactosidase-positive cells were scored for apoptotic morphology as previously described.13 As expected, DEBCL induced cell death in transfected cells (Figure 1d). BUFFY expression also resulted in robust apoptosis in most cells. Coexpression of both BUFFY and DEBCL further enhanced the cell killing effect (Figure 1d). However, the BUFFYΔMA mutant was completely nontoxic in transfected cells suggesting that the MA domain of BUFFY is essential for its proapoptotic activity, probably by targeting the protein to the correct site of action.
BUFFY contains a functional NLS
BUFFYΔMA may accumulate in the nucleus by passive diffusion or by active targeting. Analysis of the first 118 amino acids of BUFFY revealed a putative nuclear localization signal (NLS)14 comprised of basic, charged residues KRRLRR. To assess whether this putative NLS was functional, we generated several chimeric proteins and truncation mutants as illustrated in Figure 2a. Fusion of GFP to this N-terminal region (GFP-BUFFY1-118) eliminated the cytoplasmic GFP fraction and shifted GFP localization entirely to the nucleus (Figure 2b). Mutation of three of the basic residues in the putative NLS to alanine (KRRLRR to AAALRR) relocalized BUFFY into the cytoplasm with punctate staining that partially colocalized with ER-Tracker (Figure 2b). To further investigate this possibility, BUFFY1-118 was fused to the C-terminal region of the cytoplasmic caspase, STRICA. This chimeric molecule localized predominantly to the nucleus in both SL2 and BG2 cell lines (Figure2c and data not shown). Mutation of the NLS caused a shift in localization into the cytosol (Figure 2c). These findings indicate that the N-terminal region of BUFFY contains a functional NLS.
The N-terminal 118 amino acids of BUFFY can target proteins to the nucleus in SL2 cells. (a) Schematic representation of constructs used to assess the ability of the N-terminal 118 amino acids of BUFFY to target heterologous proteins to the nucleus. Full-length, BUFFY is shown at the top. The wild-type sequence, KRRLRR, was mutated to AAALRR as indicated. (b) Fusion of the N-terminal 118 amino acids of BUFFY to GFP targets the protein exclusively to the nucleus. (c) A fusion between the N-terminal 118 amino acids of BUFFY with the C-terminal region of STRICA was expressed in Drosophila SL2 cells. Mutation of the putative NLS results in diffuse localization throughout the cell (bottom row), compared with the nuclear localization in the wild-type NLS fusion (top row). (d) FLAG-BUFFYΔMA in SL2 cells localizes exclusively to the nucleus. Nuclei are stained with Hoechst. Mutation of the NLS in GFP-BUFFYΔMA (mutNLS) still has a fraction of protein present in the nucleus, but expression shifts to include the cytoplasm of the cell, demonstrating that the amino-acid changes prevent the targeting of protein to the nucleus
As shown in Figure 1c, removal of the MA (ΔMA) of BUFFY was sufficient to allow protein localization to the nucleus. Mutation of the NLS in BUFFYΔMA disrupted the nuclear localization in SL2 cells (Figure 2d). Together, these results indicate that BUFFY contains a functional NLS that is able to target BUFFY to the nucleus. While we have never detected a cleaved, ΔMA form of BUFFY in vivo, it is possible that in some contexts, post-translational modifications may result in the targeting of BUFFY to the nucleus to sequester its apoptosis-inducing role.
DEBCL, but not BUFFY, contains a MOM-targeting sequence
A study carried out by Kaufmann et al.5 identified residues within the C-terminal MA region flanking the transmembrane (TM) domain in Bcl-XL that are required for targeting Bcl-XL to the MOM. Comparison between the MOM-targeting sequence of Bcl-XL with other known mitochondrial-targeted proteins led the authors to identify a consensus for MOM targeting. This sequence is Bx0–9Bx0–2-TM-x0–1Bx0–6B where B is a basic residue such as Arg (R) or Lys (K) flanking the TM domain (Figure 3). In the absence of such a targeting sequence, TM-containing proteins distribute to other membranes including the ER.
Comparison of the C-terminal TM domains of Bcl-XL, Bcl-2, DEBCL and BUFFY. Various amino-acid differences in the TM flanking regions of Bcl-XL and Bcl-2 responsible for the differential intracellular targeting of these two Bcl-2 homologues.5 The X domain describes the region in Bcl-XL that possesses various positive charges absent in Bcl-2. The corresponding region in Bcl-2 is denoted as the X/2 domain. The basic (B) domain in Bcl- XL represents the two basic charges at the C-terminal end of the TM domain (TMB). The consensus for mitochondrial outer membrane (MOM)-targeting is shown in the boxed section. Basic residues are shown in bold type. Analysis of the amino-acid sequences in BUFFY and DEBCL show that DEBCL contains residues required for MOM-targeting while these residues are lacking in BUFFY
As shown in Figure 1, BUFFY localizes to the ER in transfected cells. In contrast, DEBCL localizes to the MOM.10 A comparative analysis of DEBCL and Bcl-XL showed that the X domain conforms to the consensus, with two positively charged arginine residues in both proteins, N-terminal to the hydrophobic TM (Figure 3). Additionally, Bcl-XL and DEBCL each possess a double positive charge at the C-terminus of the MA. Bcl-2 and BUFFY, on the other hand, lack such strong positive charges (Figure 3) allowing for localization to other intracellular membranes. Thus, the residues required for MOM-targeting are conserved between Bcl-XL and DEBCL, and are lacking in Bcl-2 and BUFFY.
Positive charges flanking the TMB of DEBCL are required for MOM-targeting
As previously reported, the minimal region required for targeting Bcl-XL to the MOM include the X domain and the TM followed by two basic amino acids (TMB)5 (Supplementary Figure 1a). In NIH-3T3 cells, GFP-X-TMB(Bcl-XL) effectively targeted to mitochondria and colocalized with cytochrome c (Supplementary Figure 1b). Of the four Arg residues in the X domain of Bcl-XL, the two closest to the TM were shown to be sufficient for MOM targeting.5 Mutations of these two basic residues in the X domain of Bcl-XL [GFP-AFNA-TMB(Bcl-XL)] resulted in loss of MOM targeting.5 However, in NIH3T3 cells, this Bcl-XL mutant did not notably mislocalize the protein, with most expressing cells still costaining with cytochrome c (Supplementary Figure 1b). In contrast, mutation of the C-terminal-most Arg residues in the B domain had a profound effect on Bcl-XL localization. In this case, mutation of either the Arg or Lys residues in Bcl-XL resulted in diffuse cytoplasmic staining and perinuclear localization (Supplementary Figure 1c). Thus, the C-terminal B domain, containing two positive charges in Bcl-XL, plays a crucial role in targeting Bcl-XL to mitochondrial membranes, whereas positive charges in the X domain have a less significant effect.
To assess the importance of these conserved basic residues on the localization of DEBCL, we generated GFP-fusion protein constructs containing mutations in the X and/or B-domain (Figure 4a). The presence of a tag at the C-terminus of MA-containing proteins is unsuitable, resulting in protein mislocalization, even with short tags such as FLAG,5 so all constructs were N-terminally tagged (Figure 4a). As expected, GFP-DEBCL colocalized with cytochrome c in NIH3T3 (Figure 4b) and with Mitotracker in SL2 cells (Figure 4c) confirming that DEBCL is targeted to the MOM. Mutation of two positively charged Arg residues in the X domain of DEBCL (GFP-DEBCL mut X) did not significantly disrupt the localization of the protein (Figure 4b and c); however, a portion of protein did appear to be more perinuclear in SL2 cells (Figure 4c). Mutation of the two C-terminal Arg residues (GFP-DEBCL RR/AA) slightly disrupted the mitochondrial localization of DEBCL and caused a portion of the protein to assume cytoplasmic localization (Figure 4b and c). Despite each of these mutant versions of DEBCL having little effect on the mitochondrial localization of the protein, mutation of both the X domain and C-terminal charges, GFP-DEBCL double mutant (DM), resulted in a more notable diffuse localization pattern with an increase in perinuclear staining, compared with the wild-type DEBCL (Figure 4b and c). Furthermore, deletion of the MA (ΔMA) caused DEBCL to become cytoplasmic with some protein detectable in the nucleus (Figure 4c). Together, these results demonstrate that both the X domain and B domain contribute to DEBCL mitochondrial localization.
Basic residues flanking the TMB contribute to MOM-targeting of DEBCL in transfected cells. (a) Various mutant DEBCL constructs used in the study. (b) NIH3T3 or (c) SL2 cells transfected with GFP-DEBCL and costained with cytochrome c antibody or MitoTracker Red demonstrate that DEBCL localizes to mitochondria (b and c, top panels). Mutation of two Arg residues to Ala in the DEBCL X domain (DEBCL mutX) did not strongly affect the localization, with most of the protein still costained with mitochondria and DEBCL RR/AA exhibits slight mislocalization with an increase in perinuclear localization and more diffuse cytoplasmic staining. DEBCL DM possesses both mutX and RR/AA mutations and has a more severe effect on protein localization. Deletion of the MA causes DEBCL localization predominantly to cytosol (c, bottom panels)
To confirm the altered localization of these DEBCL mutants, we carried out cellular fractionation experiments to separate mitochondrial, ER and cytosolic fractions.15 As shown in Figure 5, DEBCL is predominantly present in heavy membrane (HM) fractions comprising mitochondria. While DEBCL mutants could still localize to HM, there was a notable amount of protein in cytosolic fractions confirming our immunofluorescence data that mutation of the X domain and B domain can significantly mislocalize DEBCL protein (Figure 5). Truncated DEBCL (ΔMA) protein was only detected in cytosolic fractions indicating that the MA is essential for DEBCL mitochondrial localization (Figure 5).
DEBCL mutants localize to different cellular compartments. SL2 cells transfected with various DEBCL mutant constructs were fractionated for HM containing mitochondria, peroxisomes, Golgi; LM containing ER and Golgi membranes; and cytosol. While DEBCL protein associates with HM, mutation in the X or B-domain increases the cytosolic localization of DEBCL. The vertical white line indicates that the first lane in panels was derived from a different blot
Increasing C-terminal basicity in BUFFY alters its intracellular localization
Bcl-2 has been shown to localize to membranes of the nuclear envelope and endoplasmic reticulum in addition to the mitochondrial outer membrane.1, 5, 16 A construct expressing GFP fused to half of the X domain (X/2) and TMB of Bcl-2 [GFP-X/2-TMB(Bcl-2)] (Supplementary Figure 2a) was expressed in NIH3T3 cells and costained with Calnexin. As shown by previous studies,5 while GFP-X/2-TMB(Bcl-2) colocalized with Calnexin and displayed perinuclear localization consistent with the nuclear envelope, a significant proportion of expressed protein was observed in the cytoplasm in a punctate manner reminiscent of mitochondria (Supplementary Figure 2b). Replacement of the X/2 domain of Bcl-2 with the X domain of Bcl-XL disrupted the perinuclear staining observed in the wild-type, and increased the MOM-targeting of the protein (Supplementary Figure 2a and c). Perinuclear staining was also lost when the C-terminal basicity of Bcl-2 was increased by the addition of Arg and Lys residues (Supplementary Figure 2c), confirming the importance of a strong basic charge for mitochondrial localization, the lack of which allows protein localization to the nuclear envelope and ER.
To assess the importance of the residues surrounding the TM for localization of BUFFY, we mutated the residues around the TM to increase the basicity and mimic the X and B domains of DEBCL (Figure 6a). As shown in Figure 6b, GFP-BUFFY clearly colocalizes with Calnexin in transfected NIH3T3 cells. Increasing the basicity C-terminal to the MA [GFP-BUFFY(RRK)] had a slight effect on disrupting the perinuclear localization of BUFFY, and an increase in the amount of diffuse cytoplasmic protein in the cell was observed (Figure 6b). Additionally, some cells exhibited diffuse staining throughout the cell, suggesting that the ability to target to membranes was lost entirely. Insertion of the X domain of DEBCL N-terminal to the TM of BUFFY did not drastically alter protein localization; however, an increase in the punctate cytoplasmic localization was observed (Figure 6b). Similar localization pattern was observed when these BUFFY mutants were expressed in SL2 cells (Figure 6c).
Expression of BUFFY and BUFFY mutants in NIH3T3 and SL2 cells. (a) Various BUFFY mutant constructs were generated. (b) GFP-BUFFY colocalizes with Calnexin in transfected NIH-3T3 cells (top row). Increasing the basic charge at the end of the MA (EGFP-BUFFY RRK) increases the diffuse cytoplasmic localization of the protein. (b and c) Insertion of the X domain of DEBCL has a moderate effect on BUFFY localization. Perinuclear staining is preserved while punctate staining in the cytoplasm, indicative of mitochondrial localization, increases. (c) Expression of mutant proteins only partially colocalize with ER-Tracker in SL2 cells. (d) Cell fractions demonstrate BUFFY associates with HM and LM fractions, while BUFFY (RRK) and BUFFY (DX) display increased cytosolic localization
Analysis of protein localization by cellular fractionation demonstrated that BUFFY localizes to both HM and light membrane (LM) fractions indicating that overexpressed BUFFY localizes to ER as well as HM compartments such as lysosomes, peroxisomes or Golgi (Figure 6d). In contrast, BUFFY(RRK) localized predominantly to LM and cytosol, and insertion of the DEBCL X-domain causes protein to only associate with HM and cytosol (Figure 6d). These findings further substantiate the importance of the residues surrounding the TM for correct ER localization of BUFFY.
Mutation of the BH3 domain does not alter localization of DEBCL or BUFFY
The BH3 domain of Bcl-2 family of proapoptotic proteins is essential for their cell killing function and dimerization.1, 2, 3, 4, 17, 18, 19 In Bak, the BH3 domain forms the second α-helix that binds into a hydrophobic cleft formed by the BH1, BH2 and BH3 domains of Bcl-XL.17 Mutation of Leu78 in the Bak BH3 domain decreases its binding affinity for the hydrophobic cleft in Bcl-XL and reduces its ability to induce cell death.16 The hydrophobic residues that make up the BH3 domain are highly conserved and mutation of Leu146 residue at the beginning of the BH3 domain in DEBCL, similarly diminishes its ability to induce apoptosis.6 In contrast, we found that mutation of the Glu151 residue in DEBCL BH3 domain did not significantly affect its apoptosis-inducing ability (data not shown).
To assess whether localization of BUFFY or DEBCL could be affected by a mutations in their BH3 domain, we transfected the respective mutant constructs in SL2 cells and observed protein localization by immunofluorescence. As shown in Figure 7a, BUFFY BH3 mutant still colocalized with Calnexin indicating that ER and perinuclear localization was not affected. Interestingly, BH3 mutation had no effect on the proapoptotic activity of BUFFY (data not shown). Similarly, mutation of either Leu146 or Glu151 in DEBCL had no significant effect on mitochondrial localization (Figure 7b). Analysis of protein localization by cellular fractionation indicated that mutation of the conserved hydrophobic Leu146 residue of the DEBCL BH3 domain caused slight mislocalization of the protein from mitochondria to cytosol (Figure 7b). This suggests that the mitochondrial localization of DEBCL may be affected by the conformation of the protein and that disruption of BH3-domain hydrophobic interactions required for protein dimerization may affect DEBCL protein localization, which may explain the reduced ability of this mutant to induce apoptosis.6
Expression of BUFFY and DEBCL BH3 mutants in SL2 cells. (a) BUFFY BH3 mutant (GDE145AAA) was transfected into SL2 cells and costained with Calnexin, illustrating ER and perinuclear localization. (b) Expression of HA-DEBCL BH3 mutants in SL2 cells showing that L146G and E151G mutations does not disrupt mitochondrial localization. (c) Cell fractions showing that BH3 mutations do not significantly alter protein localization. The DEBCL L146G mutation shows partial cytosolic localization. The vertical white line indicates that the first lane in panels was derived from a different blot
Discussion
The mammalian Bcl-2 family has over 20 members, which possess either proapoptotic or antiapoptotic activity.1, 2, 3, 4 In contrast, the Drosophila genome possesses only two Bcl-2 family members, DEBCL and BUFFY.6, 7, 8, 9, 10, 11 The functions of Bcl-2 family members in Drosophila remains enigmatic as the inhibitor of apoptosis protein DIAP1 is the main regulator of caspase activation in the fly.20, 21 DEBCL was initially characterized as a proapoptotic protein; however, there is evidence to suggest that it also has prosurvival function in response to some death insults.6, 7, 8, 9, 10 Overexpression of BUFFY in Drosophila embryos can provide partial protection against radiation-induced apoptosis.11 More recent data have attributed both prosurvival and proapoptotic functions to BUFFY.12 The primary sequences of both BUFFY and DEBCL are most closely related to the proapoptotic mammalian Bcl-2 homologue Bok, a proapoptotic Bax-like protein.6, 7, 8, 9, 10, 11, 22 Results presented here demonstrate that BUFFY overexpression in cultured Drosophila cells induces cell death, similar to DEBCL.
Unlike DEBCL, which largely localizes to mitochondria, our data suggest that BUFFY is primarily associated with ER. The ER localization of BUFFY was dependent upon the hydrophobic MA at the C terminus of BUFFY, shared by most Bcl-2 homologues. Deletion of the MA causes dramatic mislocalization of BUFFY and renders the protein inactive in cell death assays. This observation indicates that the MA-dependent subcellular localization of BUFFY is required for its biological activity in inducing cell death. Disruption of the MA region in DEBCL leads to its mislocalization from mitochondria to cytosol and loss of its proapoptotic activity.10 Thus, like the mammalian Bcl-2 proteins, localization of fly Bcl-2 family members to specific intracellular membranes is mediated via the C-terminal MA regions and their proper localization is necessary for their function.
Interestingly, disruption of BUFFY MA region resulted in the protein accumulation in the nucleus. Notably, deletion of the MA domain in DEBCL did not result in localization of the protein to nucleus. The nuclear localization of BUFFYΔMA may be caused by passive entry of this 30 kDa protein, once it is removed from the ER membranes. However, we noted a potential NLS in the N-terminal region of BUFFY. Experiments fusing the N-terminal 118 amino acids of BUFFY, containing the putative NLS (KRRLRR), to heterologous proteins such as GFP and N-terminally truncated STRICA suggest that this region of BUFFY can facilitate nuclear transport of proteins. Mutation of the putative NLS (KRRLRR to AAALRR) resulted in localization of BUFFYΔMA or N-terminal 1-118 fused GFP or STRICA to cytosol indicating that the putative NLS is likely to be functional. Given very low levels of endogenous BUFFY that is not detectable with available antibodies, the physiological significance of nuclear BUFFY remains to be established. However, such a change in BUFFY localization may serve as a regulatory mechanism whereby post-translational modifications cause the nuclear targeting of BUFFY, perhaps sequestering it away from its site of action.
Since the MA is important for DEBCL and BUFFY localization and function, we focused on dissecting out the molecular determinants that regulate whether the MA directs the protein to mitochondria or ER. The study by Kaufmann et al.5 identified differences in the C-terminal amino-acid sequences of Bcl-XL, an MOM-targeted molecule and Bcl-2, that localizes to membranes of the ER and nuclear envelope in addition to mitochondria. Given the conservation of such residues in DEBCL and the absence of these in BUFFY, we predicted that the C-terminal region is responsible for the differential localization of BUFFY and DEBCL. Our work confirmed our prediction that specific amino acids flanking the hydrophobic trans-MA are required, in addition to the MA itself, for correct intracellular targeting. DEBCL shares similar residues with Bcl-XL and both of these proteins are targeted to the MOM. These results demonstrate that the strong positive charges at the C-terminus play an important role in MOM-targeting, whereas Arg residues in the X domain, while contributing to mitochondrial localization, play a less significant role.
Localization of Bcl-XL is consistent with the results obtained for DEBCL, although somewhat different to originally published data.5 Specifically, while mutation of the C-terminal charges has a drastic effect on Bcl-XL localization, as published, mutations in the X domain have little effect on MOM-targeting, in NIH3T3 cells (Supplementary Figure 1b). Correct mitochondrial targeting of DEBCL depends on both Arg residues in the X domain, as well as the two Arg residues in the B domain. While mutation of the X domain alone did not significantly mislocalize DEBCL, the B domain mutations (RR/AA) had a more marked effect. Most disruptive, however, was the effect of both mutations on DEBCL. Significant mislocalization was observed in DEBCL DM, with loss of the punctate cytoplasmic staining characteristic of mitochondria, favouring a more diffuse cytoplasmic staining. DEBCL DM did not colocalize with the mitochondrial protein cytochrome c or MitoTracker in transfected cells and exhibited an increase in localization around the nucleus, further demonstrating loss of normal localization. It should be noted that while the corresponding X domain mutations in Bcl-XL reportedly mislocalize, no significant difference was observed in our experiments. Although the X domain and B domain are required for the correct localization of DEBCL, other residues are likely to be required as well as the TMB itself, since the DEBCL mutants do not show a classic ER pattern of staining.
In the case of BUFFY, ER localization is somewhat disrupted by insertion of the X domain of DEBCL N-terminal to the BUFFY TMB. More pronounced, however, was the effect of increasing the basicity at the C-terminal end of the TMB from RF to RRK (Figure 6b and c) demonstrating the importance of more neutral amino acids for ER localization. While the wildtype EGFP-X/2-Bcl-2 did not completely costain with Calnexin in NIH3T3 cells, increasing the basicity at either TM flank increased the mitochondrial targeting of the molecule.
Thus, the data presented here demonstrate that C-terminal basicity following the TM domains of BUFFY and DEBCL plays a role in the differential localization of these molecules. Positive charges at the N-terminal flank also contribute to the correct intracellular localization of the proteins; however, the TMB itself and other residues are likely to be required for targeting to the MOM or other organelle membranes. As the precise in vivo functions of these proteins is not fully understood, the physiological consequences of the differential localizations of BUFFY and DEBCL remain to be established. However, irrespective of the biological activities of BUFFY and DEBCL, the data in this paper indicate that the primary determinants of localization (and presumably function) of these proteins lie in the C-terminal regions.
Materials and Methods
Constructs
pCDNA3-FLAG-Bcl-XL, pCDNA3-FLAG-Bcl-XL(RS), pCDNA3-FLAG-Bcl-XL(SK), pCDNA3-FLAG-Bcl-2, pCDNA3-FLAG-Bcl-2 (HKRK), pEGFP-X-TMB(Bcl-XL), pEGFP-AFNA-TMB(Bcl-XL), pEGFP-X/2-TMB(Bcl-2) and pEGFP-X(Bcl-XL)TMB(Bcl-2) were kindly provided by Professor Christoph Borner (Albert-Ludwigs-University of Freiburg, Germany). buffy and debcl cDNA6, 11 for both full length and ΔMA were amplified by PCR and cloned into pEGFPc2 vector (Clontech) to create pEGFP-BUFFY, pEGFP-BuffyΔMA, pEGFP-DEBCL and pEGFP-DEBCLΔMA. Mutations in the buffy and debcl cDNA sequences, including insertion mutations, were introduced by site-directed mutagenesis. BUFFY mutant BH3 contains GDE145VAA mutation; DEBCL BH3 mutants contain E151G or L146G mutation. BUFFY (mutNLS) incorporates KRR104AAA mutations, BUFFY-RRK mutant was generated by insertion of RRK residues following the TM domain at amino-acid position 285 and BUFFY (+DX) was generated by insertion of the DEBCL X domain residues RHIRPR261 ahead of the BUFFY TM region. DEBCL (mutX) comprises both R259A and R261A mutations, DEBCL (RR/AA) comprises RR290AA mutations and DEBCL (DM) is a DM consisting of both X-domain and RR/AA mutations. For cloning into the insect expression vector, pRMHa3, all full length and mutant buffy and debcl cDNAs were excised from pEGFPc2 vector by digestion with AfeI/BamHI and cloned into pRMHa3 via SmaI/BamHI to create GFP-tagged constructs for expression in Drosophila cells. FLAG-tagged buffy and HA-tagged debcl were generated by PCR amplification and cloned into the insect expression vector pIE1.4 (Novagen).
Cell culture
NIH3T3 cells were maintained in Dulbecco's modified Eagle's medium (GibcoBRL) supplemented with 10% FBS and 1% glutamine. COS-7 and HEK-293T were maintained in RPMI-1640 (Gibco-BRL) supplemented with 10% FBS. Media contained 2% sodium hydrocarbonate, 10% HEPES, 1% penicillin (Glaxo) and 1% steptomycin sulphate (Glaxo). Mammalian cell lines were grown at 37°C in a humidified incubator containing 5% CO2. The Drosophila cell lines, SL2 and BG2-c2, were maintained in Schneider's culture medium (Gibco) supplemented with 0.5% penicillin/streptomycin (Gibco), 1% glutamine and 10% FBS (Gibco) and grown at 27°C.15 BG2-c2 cells were also supplemented with 10 μg/ml insulin (Novartis).
Transfections
NIH3T3 or COS-7 cells were seeded at 2.5 × 105 cells per well in a six-well dish or at 5 × 105 in 60 mm dishes. Mammalian cells were transfected with buffy or debcl expression constructs using FuGENE 6 according to the manufacturer's protocol (Roche). Drosophila cells were transfected with buffy or debcl expression constructs using Cellfectin according to the manufacturer's protocol (Invitrogen).
Immunofluorescence
Transfected cells were washed in PBS and fixed in 4% paraformaldehyde for 15 min followed by permeabilization in 0.1% Triton X-100 for 10 min at room temperature. Where indicated, live cells were incubated with 300 nM of either ER-Tracker Red (Molecular Probes) or MitoTracker Red (Molecular Probes) in SF900 II (GIBCO) serum-free medium at 27°C for 30 min, before fixation. Cells were co-stained with 2 μg/ml of either anti-FLAG mouse antibody (SIGMA) or anti-HA rat antibody (Roche). Anti-cytochrome c mouse antibody clone 6H2.B4 (Pharmingen) was used at 3 μg/ml and anti-Calnexin rabbit antibody used at 1:500 dilution (Stressgen). All antibodies were diluted in PBS with 10% FBS and incubated with cells overnight at 4°C. Cells were washed in PBS and incubated with with Alexa Fluor 488 (green), Alexa Fluor 568 (red) conjugated anti-mouse anti-rabbit or goat anti-rat secondary antibodies (Molecular Probes). For staining nuclei, Hoechst was added at 0.4 μg/ml and incubated with cells for 5 min. Fluorescence images were captured either using a confocal microscope (Radiance 2100, Biorad) or fluorescence microscope (model BX51; Olympus) and a camera (model U-CMAD3/CVM300; Olympus). Cells were visualized under × 100 UPLAPO objective lens with NA=1.3, under oil immersion and images processed with Olysia BioReport software (Olympus). All green and red fluorescent images were equally adjusted for brightness and contrast and were merged manually using Adobe Photoshop software.
Cell fractionation
5 × 106 cells were lysed in Buffer A (20 mM HEPES-KOH pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT in 250 mM sucrose supplemented with protease inhibitors) and harvested by differential centrifugation for HM LM and cytosolic fractions as previously described.15
Cell death assays
SL2 cells were transiently transfected with 1.5 μg buffy and/or debcl expression constructs together with 0.5 μg pIE1.4-LacZ reporter construct using Cellfectin. 24 h following transfection, cells were fixed in 2% formaldehyde/0.2% glutaraldehyde in PBS for 5 min. β-Galactosidase activity was visualized by staining cells with X-gal solution (1 mg/ml X-gal, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2 in PBS) for 3 h at 37°C.13 Percent survival was calculated by scoring the number of β-gal positive cells in an equal number of fields of view, compared to vector control. Assays were performed in triplicate and error bars represent ±S.E.M.
Abbreviations
- ER:
-
endoplasmic reticula
- MA:
-
membrane anchor
- NLS:
-
nuclear localization signal
- BH:
-
Bcl-2 homology
- TM:
-
transmembrane
References
Cory S, Adams JM . The Bcl-2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2002; 2: 647–656.
Schwartz PS, Hockenbery DM . Bcl-2-related survival proteins. Cell Death Differ 2006; 13: 1250–1255.
Reed JC . Proapoptotic multidomain Bcl-2/Bax-family proteins: mechanisms, physiological roles, and therapeutic opportunities. Cell Death Differ 2006; 13: 1378–1386.
Labi V, Erlacher M, Kiessling S, Villunger A . BH3-only proteins in cell death initiation, malignant disease and anticancer therapy. Cell Death Differ 2006; 13: 1325–1338.
Kaufmann T, Schlipf S, Sanz J, Neubert K, Stein R, Borner C . Characterization of the signal that directs Bcl-XL, but not Bcl-2, to the mitochondrial outer membrane. J Cell Biol 2003; 160: 53–64.
Colussi PA, Quinn LM, Huang DC, Coombe M, Read SH, Richardson H et al. Debcl, a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J Cell Biol 2000; 148: 703–714.
Igaki T, Kanuka H, Inohara N, Sawamoto K, Nunez G, Okano H et al. Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc Natl Acad Sci USA 2000; 97: 662–667.
Zhang H, Huang Q, Ke N, Matsuyama S, Hammock B, Godzik A et al. Drosophila pro-apoptotic Bcl-2/Bax homologue reveals evolutionary conservation of cell death mechanisms. J Biol Chem 2000; 275: 27303–27306.
Brachmann CB, Jassim OW, Wachsmuth BD, Cagan RL . The Drosophila bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr Biol 2000; 10: 547–550.
Igaki T, Miura M . Role of Bcl-2 family members in invertebrates. Biochim Biophys Acta 2004; 1644: 73–81.
Quinn L, Coombe M, Mills K, Daish T, Colussi P, Kumar S et al. Buffy, a Drosophila Bcl-2 protein, has anti-apoptotic and cell cycle inhibitory functions. EMBO J 2003; 22: 3568–3579.
Senoo-Matsuda N, Igaki T, Miura M . Bax-like protein Drob-1 protects neurons from expanded polyglutamine-induced toxicity in Drosophila. EMBO J 2005; 24: 2700–2713.
Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA . Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev 1994; 8: 1613–1626.
Pemberton LF, Paschal BM . Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 2005; 6: 187–198.
Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S . The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol 2002; 156: 1089–1098.
Lithgow T, van Driel R, Bertram JF, Strasser A . The protein product of the oncogene bcl-2 is a component of the nuclear envelope, the endoplasmic reticulum, and the outer mitochondrial membrane. Cell Growth Differ 1994; 5: 411–417.
Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M et al. Structure of Bcl-xL–Bak peptide complex: recognition between regulators of apoptosis. Science 1997; 275: 983–986.
Wang K, Gross A, Waksman G, Korsmeyer SJ . Mutagenesis of the BH3 domain of BAX identifies residues critical for dimerization and killing. Mol Cell Biol 1998; 18: 6083–6089.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ et al. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 2001; 292: 727–730.
Kumar S . Caspase function in programmed cell death. Cell Death Differ 2007; 14: 32–43.
Hay BA, Guo M . Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol 2006; 22: 623–650.
Zhang H, Holzgreve W, De Geyter C . Evolutionarily conserved Bok proteins in the Bcl-2 family. FEBS Lett 2000; 480: 311–313.
Acknowledgements
We are grateful to Christoph Borner for providing the Bcl-XL and Bcl-2 constructs used in this study. We thank the members of our laboratory for helpful comments. This work was supported by the National Health and Medical Council of Australia. JD was supported by a Royal Adelaide Hospital Dawes Postgraduate Award. LD is a Royal Adelaide Hospital Florey Fellow.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Doumanis, J., Dorstyn, L. & Kumar, S. Molecular determinants of the subcellular localization of the Drosophila Bcl-2 homologues DEBCL and BUFFY. Cell Death Differ 14, 907–915 (2007). https://doi.org/10.1038/sj.cdd.4402082
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4402082
Keywords
This article is cited by
-
Apoptosis in Drosophila: which role for mitochondria?
Apoptosis (2016)
-
The Bcl-2 family: structures, interactions and targets for drug discovery
Apoptosis (2015)
-
Drosophila larvae lacking the bcl-2 gene, buffy, are sensitive to nutrient stress, maintain increased basal target of rapamycin (Tor) signaling and exhibit characteristics of altered basal energy metabolism
BMC Biology (2012)
-
The zebrafish as a model organism for the study of apoptosis
Apoptosis (2010)
-
Cellular responses to endoplasmic reticulum stress and apoptosis
Apoptosis (2009)