Abstract
Breast cancer cells often show increased activity of the mitogen-activated protein kinase (MAPK) pathway. We report here that this pathway reduces their sensitivity to death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and present the underlying mechanism. Activation of protein kinase C (PKC) inhibited TRAIL-induced apoptosis in a protein synthesis-independent manner. Deliberate activation of MAPK was also inhibitory. In digitonin-permeabilized cells, PKC activation interfered with the capacity of recombinant truncated (t)Bid to release cytochrome c from mitochondria. MAPK activation did not affect TRAIL or tumor necrosis factor (TNF)α-induced Bid cleavage. However, it did inhibit translocation of (t)Bid to mitochondria as determined both by subcellular fractionation analysis and confocal microscopy. Steady state tBid mitochondrial localization was prohibited by activation of the MAPK pathway, also when the Bcl-2 homology domain 3 (BH3) domain of tBid was disrupted. We conclude that the MAPK pathway inhibits TRAIL-induced apoptosis in MCF-7 cells by prohibiting anchoring of tBid to the mitochondrial membrane. This anchoring is independent of its interaction with resident Bcl-2 family members.
Similar content being viewed by others
Introduction
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is one of the several members of the tumor necrosis factor (TNF) gene superfamily that induce apoptosis through engagement of death receptors.1 TRAIL selectively induces apoptosis in many transformed cells but is relatively nontoxic to normal cells.2 Upon binding to its proapoptotic receptors TRAIL induces the formation of the death-inducing signaling complex (DISC) by recruitment of the dual adaptor molecule Fas-associated death domain (FADD) through its death domain (DD) that in turn, recruits the initiator caspase-8 through its death effector domain (DED).3 Caspase-8 is then activated at the DISC through oligomerization. Active caspase-8 can process Bcl-2 family member Bid to kill the cell by a mitochondrial-regulated apoptotic pathway.3 Active caspase-8 can also directly activate executioner caspases to induce cell death by a mitochondria-independent pathway.4 In the mitochondria-dependent apoptotic pathway, caspase-8-mediated cleavage of Bid generates a truncated form of Bid (tBid) that translocates to the mitochondria and promotes the release of apoptogenic factors in concert with proapoptotic Bax and Bak proteins.5, 6, 7 Cytosolic cytochrome c (cyt c) enables apoptosome formation (a complex formed by Apaf-1, cyt c, dATP and caspase-9).8 Apoptosome formation leads to caspase-9 activation which in turn processes and activates the executioner caspases.
Protein kinase C (PKC) counteracts apoptosis-induction by various stimuli,9, 10 including death receptors.11, 12 One of the downstream effectors of PKC antiapoptotic action is the mitogen-activated protein kinase (MAPK) pathway13 although MAPK-independent mechanisms may also be operative.12 The PKC-MAPK pathway has been shown to inhibit TRAIL-induced apoptosis by various investigators. However, the mechanism by which this is achieved is unclear. Activation of PKC has been suggested to inhibit TRAIL-induced apoptosis by preventing FADD recruitment to the DISC14, 15 or reducing the processing of procaspase-8.16 On the other hand, it was reported that an activated MAPK pathway abrogated Smac/DIABLO release from mitochondria.17 Our previous work indicated that the PKC-MAPK pathway impacted on the mitochondrial route of effector caspase activation.18
The present study has explored this possibility and pinpointed the inhibitory effect of the MAPK pathway upon TRAIL induced apoptosis. We find that active MAPK can confer resistance to TRAIL-induced apoptosis by disrupting the association of tBid with the mitochondrial membrane. This does not involve dissociation of its Bcl-2 homology domain 3 (BH3)-domain-mediated interaction with Bcl-2 family members, but interference with anchoring to other membrane-associated components.
Results
Protein synthesis-independent inhibition of TRAIL-induced apoptosis by PKC-mediated MAPK activation in breast tumor MCF-7 cells
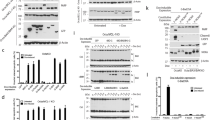
PKC-mediated MAPK activation has previously been reported to inhibit TRAIL-induced apoptosis.14, 15, 16, 17, 18 Accordingly, it is well known that MAPK activates genes implicated in cell survival.19 To determine whether inhibition of TRAIL-induced apoptosis exerted by PKC activation is due to the novo synthesis of survival genes, MCF-7 cells were treated with TRAIL and PKC activator phorbol-12,13-dibutyrate (PDBu) in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX) (Figure 1a). Apoptosis was determined as PS exposure since MCF-7 cells lack caspase-3 which is required for DNA fragmentation and morphological changes of apoptosis.20 CHX did not interfere with PDBu-induced protection, indicating that the apoptosis inhibitory effect is independent of new protein synthesis. Moreover, analysis of viable cells indicated that PKC activation resulted in the inhibition of TRAIL-induced cell death both in the absence and presence of CHX (Figure 1b). To further confirm the inhibitory effect of PDBu on TRAIL-induced apoptosis, we determined the proteolytic degradation of the caspases substrate poly(ADP-ribose)polymerase (PARP). As shown in Figure 1c, PARP cleavage induced by TRAIL was clearly prevented in MCF-7 cells upon treatment with the PKC activator.
The PKC-MAPK pathway does not require de novo protein synthesis to antagonize TRAIL-mediated apoptosis. (a) MCF-7 cells were pre-treated with PDBu (20 ng/ml) for 1 h before adding TRAIL (250 ng/ml) and incubated for 6 h, in the presence or not of cycloheximide (CHX) (5 μg/ml). CHX was added 1 h before PDBu. Apoptosis was assessed by flow cytometry after staining the cells with Annexin V and propidium iodide. Error bars represent S.D. from three independent experiments. UT=untreated cells. (b) MCF-7 cells were pretreated with PDBu (20 ng/ml) for 1 h before adding TRAIL (250 ng/ml) and incubated for 24 h, in the presence or not of cycloheximide (CHX) (5 μg/ml). CHX was added 1 h before PDBu. Viable cells were determined by the crystal violet method. Results show the average and range of two independent experiments. (c) Cells were treated as in (a) and then PARP cleavage was detected by immunoblotting with anti-PARP antibody. (d) Cells were transfected by electroporation with a constitutively activated MEK1 (CA-MEK1) construct and treated with TRAIL (100 ng/ml) or TNFα (10 ng/ml) plus CHX (500 ng/ml) for 15 h. Apoptosis was assessed by measuring PS externalization by flow cytometry. Error bars represent S.D. from three independent experiments. (e) Erk1/2 activation of cells treated with PDBu for 6 h or electroporated with CA-MEK1 was assessed by immunoblotting with anti-phospho Erk1/2 antibody. The asterisk indicates a background band that serves as a loading control
A constitutively active form of the MAPK activator MEK-1 (CA-MEK1) also rendered the cells insensitive to apoptosis induced by TRAIL or TNF-α (Figure 1d). Mock transfections with empty vector did not affect the number of apoptotic cells. Activation of the MAPK pathway was verified by measuring the phosphorylation of the Erk1/2 MAP kinases (Figure 1e). Transfection of either CA-MEK1 or incubation of MCF-7 cells with PDBu induced an increase in the amount of phosphorylated Erk1/2 (Figure 1e). These findings indicate that PKC activation as well as MAPK activation by MEK inhibits TRAIL-induced apoptosis in MCF-7 cells.
Inhibition impacts on the mitochondrial route of caspase activation and selectively affects death receptor signaling
MCF-7 cells, which are deficient in caspase-3 expression,20 have been described as belonging to the type II group of cells that require the mitochondrial pathway for death receptor-mediated apoptosis signaling.4 Moreover, we have reported that overexpression of Bcl-2 in MCF-7 cells blocks apoptosis induced by CD95 antibody21 and TRAIL.22 However, reconstitution of MCF-7 cells with caspase-3 converts them to type I cells in which mitochondrial permeabilization is not required for effector caspase activation and consequent apoptosis.4 To gain further insight into the mechanism of apoptosis inhibition by the PKC-MAPK pathway, we assessed the effect of PDBu on TRAIL-induced apoptosis in caspase-3-proficient MCF-7 cells (MCF-7caspase−3). In these cells, pre-incubation with PDBu did not inhibit TRAIL-induced apoptosis (Figure 2a). These results indicate that the PKC-MAPK pathway inhibits TRAIL-induced apoptosis at some step in the mitochondrial route of effector caspase activation.
Selective inhibition of TRAIL-induced mitochondria-operated apoptotic pathway by phorbol ester. (a) MCF-7 cells overexpressing caspase-3 were treated with PDBu (20 ng/ml) for 1 h before adding TRAIL (100 ng/ml) for 5 h. Apoptosis was assessed by measuring PS externalization by flow cytometry. (b) Cells were treated with doxorubicin (DXR) (500 ng/ml) for 48 h and cell viability was determined by the crystal violet method. Some cultures were incubated with PDBu (20 ng/ml) for 1 h prior to the addition of DXR. Error bars represent S.D. from three independent experiments
It has been demonstrated that in MCF-7 cells, doxorubicin induces apoptosis through the intrinsic mitochondrial pathway.22 Furthermore, in both death receptor and genotoxic damage-induced mitochondria-operated apoptosis, proapoptotic Bcl-2 family members Bax and Bak are involved.6 To further investigate the mechanism by which the PKC-MAPK pathway inhibits TRAIL receptor-mediated apoptosis in MCF-7 cells, we examined the effect of PDBu on doxorubicin-induced apoptosis. As shown in Figure 2b, treatment with PDBu did not inhibit cell death by doxorubicin, suggesting that inhibition impacts on a signaling component that is uniquely involved in the death receptor pathway, and not on, for example, Bax or Bak.
The PKC-MAPK pathway prohibits truncated Bid from permeabilizing mitochondria
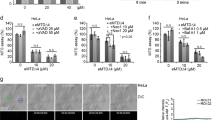
Since truncated Bid is the key component that links death receptors to the mitochondrial route of effector caspase activation, we tested the impact of the PKC-MAPK pathway on endogenous (t)Bid formation and function. As previously reported,18 TRAIL-induced processing of apical procaspase-8 was not affected by treatment with PDBu (Figure 3a). Regarding Bid processing we observed that disappearance of full length Bid was not inhibited by PKC-MAPK activation, although we always observed a smaller amount of tBid in cultures treated with PDBu. In order to test whether (t)Bid translocation to the pellet fraction containing mitochondria was affected by activation of the PKC-MAPK pathway, we performed subcellular fractionation experiments in MCF-7 cells treated with TRAIL. As shown in Figure 3b, TRAIL induced the localization of (t)Bid to the mitochondrial fraction. Interestingly, activation of the PKC-MAPK pathway prevented TRAIL-induced (t)Bid association with the mitochondrial fraction (Figure 3b). However, inhibition of tBid association with mitochondria did not result in its presence in the cytosolic fraction. This is in accordance with the results shown in Figure 3a where the amount of truncated Bid in samples from cells treated with TRAIL and PDBu was significantly reduced when compared to samples from TRAIL-treated cells despite similar full-length Bid processing. These findings suggest that cytosolic tBid is an unstable protein that can be subjected to proteolytic degradation under conditions that prevent its mitochondrial localization. Inhibition by the PKC-MAPK pathway of TRAIL-induced tBid translocation to mitochondria resulted in a marked reduction of cyt c release from this organelle (Figure 3c). To get further evidence that a step downstream (t)Bid formation could be the target of an activated PKC-MAPK pathway, we carried out a number of experiments with recombinant (t)Bid in digitonin-permeabilized cells. Addition of recombinant tBid to digitonin-permeabilized MCF-7 cells caused a marked release of cyt c from the mitochondrial fraction isolated from control cells (Figure 3d). However, in cells that had been pretreated with PDBu, tBid was unable to release cyt c from mitochondria (Figure 3d). Altogether, these data demonstrate that the PKC-MAPK pathway inhibits tBid function.
MAPK inhibits TRAIL-induced mitochondria-operated apoptotic pathway downstream of Bid cleavage and upstream of cytochrome c release. (a) MCF-7 cells were incubated in absence or presence of 20 ng/ml PDBu for 1 h and subsequently treated with or without 250 ng/ml TRAIL for 3 h. Caspase-8 processing was determined by Western blot using mouse anti-human caspase-8mAb. Both the 55±53 kDa native forms and the 43±41 kDa intermediate cleavage products are shown by arrows. The p22 full length Bid and the caspase-cleaved p15 were determined by Western blot using rabbit anti-Bid polyclonal antibody. For Bid, bands shown in upper panel were revealed for a shorter period than those shown in lower panel. (b) Cells were treated with or without PD 98059 (50 μM) for 1 h. Then the cells were incubated with or without PDBu (20 ng/ml) for 45 min before adding TRAIL (500 ng/ml) for 4 h. Levels of Bid and tBid (revealed for a longer period than Bid) in cytosolic and pellet fractions were determined by Western blot. Following treatment, cells were lysed and cytosolic proteins were separated from mitochondria as described under Materials and Methods. COX-IV was used as a mitochondrial marker and loading control. The asterisk indicates a background band that serves as a loading control. (c) Cells were treated as in (b) and levels of cytochrome c in cytosolic and membrane fractions were determined by Western blot. The asterisk indicates a background band that serves as a loading control. (d) Cells incubated with or without PDBu (20 ng/ml) were permeabilized for 10 min with a buffer containing digitonin (100 μg/ml) in the presence or not of 500 nM recombinant tBid. Cells were collected by centrifugation and supernatant or pellet proteins were resolved by immunoblotting with anti-cytochrome c antibody as described in materials and methods. Results are representative of at least three different experiments
MAPK activation inhibits TRAIL and TNFα-induced translocation of tBid to mitochondria
To examine how the PKC-MAPK pathway impacted on Bid function in intact cells, we ectopically expressed Bid-green fluorescent protein (GFP) chimeric proteins (Figure 4a) in MCF-7 cells and examined their subcellular localization by confocal microscopy. Firstly, MCF7 cells were transiently transfected with full-length (FL) Bid-GFP fusion protein and treated with TRAIL or TNFα. In untreated cells, Bid-GFP showed a diffuse cytosolic distribution (Figure 4b). After 3 h stimulation with TRAIL or TNFα, Bid-GFP fluorescence showed a punctate distribution indicative of mitochondrial localization. This was confirmed by colocalization with the mitochondrial dye MitoTracker-Red. When the transfected cells were incubated with PDBu prior to treatment with death ligand, a fraction of cells displayed cytosolic Bid-GFP distribution similar to untreated cells. Pretreatment with the specific MEK1 inhibitor, PD98059, abolished the inhibition by PDBu of death ligand-induced Bid-GFP translocation (Figure 4b). Incubation of MCF-7 cells with either PD98059 or PDBu alone had no effect on Bid-GFP localization. These effects on Bid-GFP translocation were quantified by counting cells that showed a punctate green fluorescent pattern (Figure 4c). In parallel, we showed by immunoblotting (using an antibody that recognizes GFP) that PDBu did not affect the FL-Bid-GFP cleavage (Figure 4d). Together, these results suggest that death receptor-induced translocation of Bid-GFP to mitochondria is inhibited by an activated MAPK pathway at a step downstream of caspase 8 mediated Bid cleavage.
Activation of the MAPK pathway inhibits TRAIL- or TNF-induced tBid mitochondrial localization. (a) GFP constructs of Bid and two different truncated forms of Bid. (b) MCF-7 cells were transiently transfected with full-length Bid-GFP (FL Bid) using the FuGENE 6 transfection reagent and treated with PD98059 (50 μM) for 1 h before adding PDBu (20 ng/ml) for 30 min and then treated with TRAIL (100 ng/ml) or TNFα (10 ng/ml) for a further 3-h period. At 1 h before the TNFα treatment, cells were pretreated with CHX (500 ng/ml). Cells were fixed and analyzed by confocal microscopy. Mitochondria were identified with Mitotracker Red and nuclei with ToPro-3 (blue). (c) Quantification of mitochondria localized Bid by counting cells with a punctate green fluorescent pattern as described in the Materials and Methods section. Error bars represent S.D. from three independent experiments. (d) Full-length Bid-GFP cleavage was determined by Western blot of cytosolic proteins with anti-GFP antibody
Localization of truncated Bid at the mitochondria is reversed in the presence of an activator of the MAPK pathway
We next addressed whether the MAPK pathway interfered with mitochondrial translocation of tBid or with its mitochondrial association. For this purpose, we transfected MCF-7 cells with a GFP-tBid construct. In untreated cells, GFP-tBid fluorescence was punctate and colocalized with MitoTracker, indicative of mitochondrial localization in a majority of transfected cells (Figure 5a and b). Interestingly, treatment with PDBu caused a reduction in the number of cells showing a mitochondrial localization of GFP-tBid. An activated MAPK pathway was responsible for the observed inhibition as the effect of PDBu was reversed in the presence of PD98059 (Figure 5a and b). A constitutively active form of PKC-theta, a known activator of the MAPK pathway,23 also inhibited the mitochondrial localization of GFP-tBid (data not shown).
Activation of the MAPK pathway reduces the mitochondrial localization of two different truncated forms of Bid. MCF-7 cells transiently transfected either with GFP-tBid (a and b) or GFP-ttBid (c and d) using the FuGENE 6 transfection reagent were treated with PD98059 (50 μM) (PD) for 1 h before adding PDBu (20 ng/ml) for 4 h. Cells were fixed and analyzed by confocal microscopy. Mitochondria were identified with Mitotracker Red and nuclei with ToPro3 (blue). Quantitative analysis of the mitochondrial localization of either GFP-tBid (b) or GFP-ttBid (d) by counting cells with a punctate green fluorescent pattern. Error bars represent S.D. for three independent experiments. (e) MCF-7 cells transiently transfected with GFP-tBid were treated with CHX 1 h before the addition of PDBu (20 ng/ml). Cells were then incubated for 6 h and GFP-tBid localization was assessed by confocal microscopy. Quantification of mitochondria localized GFP-tBid was performed by counting cells with a punctate green fluorescent pattern. Results show the average and range of two independent experiments
We next determined whether MAPK activity inhibited tBid mitochondrial association by disrupting tBid interactions with mitochondrial Bcl-2 family members. MCF-7 cells were transfected with a different tBid construct, GFP-ttBid, with a disrupted/nonfunctional BH3 domain (Figure 4a). The BH3 domain is essential for the interaction with other Bcl-2 proteins.24 As shown in Figure 5c and d, GFP-ttBid also localized to the mitochondria and this was partially inhibited by PDBu. Again, PD98059 reversed the PDBu effect (Figure 5d). These data indicate that the MAPK pathway does not disrupt tBid localization at the mitochondrial membrane by interfering with its interaction with other Bcl-2 family members.
To further substantiate that the MAPK pathway reversed tBid localization at the mitochondria, MCF-7 cells were transiently transfected with GFP-tBid and 15 h later treated with PDBu in the presence of cycloheximide to prevent new synthesis of GFP-tBid. Cells where then fixed and GFP-tBid localization was evaluated by confocal microscopy. At time 0 h, before addition of PDBu and cycloheximide, almost all cells show a mitochondrial localization of GFP-tBid. Interestingly, the number of cells with a mitochondrial GFP-tBid localization decreased in the presence of PDBu (Figure 5e). These results suggest that the MAPK pathway reverses the localization of tBid from the mitochondria to the cytosolic compartment.
Discussion
Despite the potential of TRAIL as an antitumor therapeutic treatment, little is known about the mechanisms regulating the sensitivity of tumor cells to TRAIL-induced apoptosis. Expression of TRAIL decoy receptors was suggested to contribute to TRAIL resistance.25, 26 However, evidence for this proposition have been based mainly in overexpression studies.25, 26 So far, there is no clear correlation between expression of these receptors and sensitivity to TRAIL-induced apoptosis in different tumor cell lines.27 Conversely, expression of proapoptotic TRAIL receptors is controlled by a variety of signals and may determine the sensitivity of tumor cells to TRAIL.28 Nevertheless, available evidence indicates that in most cases regulation of tumor cell sensitivity to TRAIL relies on intracellular mediators. Despite the fact that many cancer cells are sensitive to TRAIL-induced apoptosis, a number of them, and in particular, breast cancer cells, are resistant to this death-ligand.29
Activity and expression of MAPK are significantly elevated in breast carcinomas.30 Furthermore, it has been reported that Bcl-2 expression is upregulated by Erk31 and that the MAPK pathway could antagonize death receptor-mediated apoptosis by inducing FLICE-inhibitory protein (cFLIP) expression.32 However, our results indicate that the apoptosis antagonizing effect of a PKC/MAPK activator does not require de novo protein synthesis. Several authors have shown that MAPK pathway protects from apoptosis triggered by different stimuli at the level of mitochondria.33, 34 MCF-7 cells, which are deficient in caspase-3 expression,20 have been ascribed to the type II group of cells since they require the mitochondrial pathway for death receptor-mediated apoptosis signaling.4 Moreover, we have observed that overexpression of Bcl-2 in MCF-7 cells not only blocks CD95 mediated apoptosis21 but also TRAIL-mediated cell death.22 However, overexpression of caspase-3 converts MCF-7 cells to type I cells in which activation of executioner caspases is not regulated through the mitochondria.4 Our data show that PDBu could no longer protect MCF-7 cells overexpressing caspase-3 from TRAIL-induced apoptosis, further supporting the idea that an activated MAPK pathway inhibits TRAIL-induced apoptosis via the mitochondrial pathway.
Previous reports have suggested that in HeLa and Jurkat T cells inhibition of TRAIL-induced apoptosis by MAPK activation occurs by modulating DISC activity.14, 15, 16, 35 In contrast, in melanoma and breast cancer cell lines signaling through the MAPK/Erk pathway inhibits TRAIL-induced translocation of Bax from the cytosol to mitochondria thus preventing the release of apoptogenic factors from this organelle.17, 18 In the present report, we have demonstrated that in MCF-7 breast cancer cells TRAIL and TNF-α-induced cleavage of Bid was unaffected by PKC activation and identified the mitochondrial localization of tBid as a novel target of MAPK mediated apoptosis inhibition. How this occurs is unknown at present. One model is that MAPK activity inhibits tBid binding to Bcl-2 family proteins which is dependent upon the BH3 domain of tBid.36, 37, 38 However, our results with the GFP-ttBid construct that lacks the BH3 domain but is still subject to MAPK-mediated inhibition rule out this possibility. Alternatively, it has been described that myristoylation of tBid is required for its translocation to mitochondria.39 However, it is unlikely that an alteration of this post-translational modification by PKC/MAPK could be responsible for the inhibition of the mitochondrial localization of truncated Bid as the GFP-ttBid construct lacks the specific myristoylation sequence (also GFP-tBid can not be myristoylated since the GFP blocks the glycine target site).
Bid can be phosphorylated by casein kinases I and II leading to resistance to cleavage by caspase-8.40 However, phosphorylation of tBid has not been reported and Bid does not seem to contain any potential consensus sequences for kinases like the mitogen-activated protein (MAP) kinases Erk1/2 or Akt. On the other hand, caspase-8 cleaved Bid (tBid) can be rapidly degraded by the proteasome.41 In this respect, it is interesting that Bim(EL), another ‘BH3-only’ protein, is phosphorylated following the activation of the Erk1/2 pathway and this leads to a substantial degradation of BimEL via the proteasome pathway.42 Whether tBid stability could be altered indirectly by MAPK/Erk activation remains to be elucidated.
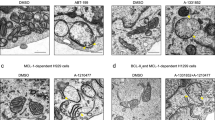
Our results with permeabilized cells may suggest an additional mechanism for the observed PKC/MAPK-mediated inhibition of mitochondrial tBid localization and TRAIL-induced apoptosis. In these experiments, cells were pretreated with a PKC activator before incubating them with recombinant tBid for a short period. Under these conditions cyt c release was markedly inhibited. These observations may indicate that PKC/MAPK activation could function to maintain mitochondrial integrity as recently reported for Akt,43 though the induction of hexokinase association with mitochondria. This association prevents the proapoptotic effect of certain members of the Bcl-2 family by preventing their mitochondrial localization.44 We are currently investigating this hypothesis in order to get further insight into the mechanism underlying the PKC/MAPK-mediated abrogation of TRAIL and TNF-α-induced apoptosis, particularly in those cells where inhibition occurs downstream of Bid cleavage by caspase-8. These studies may also help in the elucidation of the molecular mechanisms controlling tBid binding to the mitochondrial membrane.
Materials and Methods
Reagents and antibodies
RPMI 1640 medium and fetal bovine serum were obtained from Life Technologies-Europe. Soluble human recombinant TRAIL was either purchased from Alexis Biochemicals (Lausanne, Switzerland) or generated as His-tagged recombinant TRAIL in our laboratory as previously described.45 TNF alpha was from Peprotech EC LTD (London, UK). Doxorubicin (DXR), propidium iodide (PI) and phorbol-12,13-dibutyrate (PDBu) were from Sigma. (Poole, UK). PD98059 was purchased from Calbiochem-Novabiochem GmbH (Band Soden, Germany). Rabbit anti-Bid polyclonal antibody was generously provided by Dr. X Wang (Howard Hughes Medical Institute, Dallas, Texas). Mouse anti-cyt c mAb was from Pharmigen (San Diego, CA). Mouse anti-phospho-Erk1/2 mAb recognizing activated Erk1/2 was from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal anti-cyt c oxidase polypeptide IV (Cox IV) was purchased from Abcam (Cambridge, UK). Mouse anticaspase 8 mAb was from BD Pharmingen (Erembodegem, Belgium). Anti-poly(ADP-ribose) polymerase (PARP) polyclonal antibody was from Roche Molecular Biochemicals (Germany). Monoclonal antibody to alpha-tubulin was purchased from Sigma (Poole, UK). Anti-GFP antibody was from Molecular Probes (Leiden, The Netherlands).
Cell lines
The human tumor cell line MCF-7 was maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 40 μg/ml gentamycin at 37°C in a humidified 5% CO2, 95% air incubator. MCF-7 cells stably transfected with caspase-3 were generously provided by Dr. DR Green (San Diego, CA, USA).
Analysis of cell viability and apoptosis
Cell viability was determined by the crystal violet method as described.21 Apoptosis was assessed by flow cytometry after staining with Anexin-V-FLUOS (Roche Molecular Biochemicals) and propidium iodide by measuring phosphatidylserine (PS) exposure on the surface of apoptotic cells (propidium iodide negative). Flow cytometry was performed on a FACScan cytometer using Cell Quest software (Becton Dickinson, Mountain View, CA, USA). Apoptosis was also determined by analyzing the cleavage of the caspase substrate poly(ADP-ribose) polymerase by Western blotting with PARP antibody.
Immunoblot detection of proteins
After detachment with trypsin, cells (3 × 105) were washed with phosphate-buffered saline (PBS), and protein content was measured following cell lysis using the Bradford reagent (Bio-Rad Laboratories, USA) before adding Laemmli sample buffer. Samples were sonicated, and proteins were resolved on SDS-polyacrylamide gels and detected as described previously.21
Cellular fractionation
Following treatment, cells were lysed and cytosolic proteins were separated from mitochondria as previously described.35 Briefly, cells (7 × 105) were washed with PBS and lysed in 30 μl ice-cold lysis buffer (80 mM KCl, 250 mM sucrose, 500 μg/ml digitonin and proteases inhibitors in PBS). Then, cell lysates were centrifugated for 5 min at 10 000 × g. Proteins from the supernatant (cytosolic fraction) and pellet (membrane fraction) were mixed with Laemmli buffer and resolved on SDS-15% PAGE minigels.
Plasmid constructs
FL-Bid-GFP: Plasmid expressing the fusion protein full-length Bid (corresponding to 1-195 aa)/EGFP. The human FL-Bid cDNA was obtained from EST 286-f10 (MRC UK HGMP, Cambridge, UK) by PCR using the following primers, forward 5′-ATAAGATCTATGGACTGTGAGGTCAACA-3′ and reverse 5′-CCCGGATCCGTCCATCCCATTTCTGGCT-3′. The PCR product was cloned initially into the pEGFP-N3 vector (Clontech, BD-Biosciences, Belgium). Next, the FL-Bid-GFP fragment was excised from pEGFP-N3 vector with BglII and NotI and subcloned into the pEF4/Myc-His B vector (Invitrogen, The Netherlands). GFP-tBid: plasmid expressing the fusion protein truncated Bid (corresponding to aa 61-195) /EGFP. The human truncated Bid cDNA was obtained from EST 286-f10 by PCR using the following primers, forward 5′-ATAGTCGACGGCAACCGCAGCAGCC-3′ and reverse 5′-CCCGGATCCGTCCATCCCATTTCTGGCT-3′. The PCR product was cloned into the pEGFP-BOS plasmid, a modified version of pEGFP-C1 (Clontech, BD-Biosciences, Belgium) plasmid with the CMV promoter removed and replaced for the pEF-BOS promoter. GFP-ttBid: plasmid expressing the fusion protein ttBid (a shorter version of human truncated Bid corresponding to aa 95-169)/EGFP. This protein has a truncated and thereby non-functional BH3 domain. The ttBid cDNA was obtained from EST 286-f10 by PCR using the following primers, forward 5′-ATAAGATCTAGCATGGACCGTAGCATCC-3′ and reverse 5′-CCCGTCGACATCACGGAGCAAGGAC-3′. The PCR product was cloned directly into pEGFP-C1 (Clontech, BD-Biosciences, Belgium). CA-MEK1: The cDNA coding for a constitutively active mutant of MAP kinase kinase (MEK1) was as described.46
Cell transfections
For confocal analysis of Bid-GFP constructs transient transfection experiments were conducted in MCF-7 cells with the FuGENE 6 transfection reagent (Roche Applied Science) as indicated by the manufacturer. Briefly, 1.5 × 105 cells/well were seeded into six-well plates the day before transfection, 1 μg of full-length (FL) Bid-GFP, GFP-tBid or GFP-ttBid plasmids were mixed with the FuGENE 6 reagent and added to each well. At 15 h after transfection, cells were treated as indicated in the figure legend. In order to obtain high transfection efficiency (⩾90%) in experiments with the constitutively active mutant of MEK1 transfections were performed by electroporation as described previously.47 Briefly, 1 × 106 MCF-7 cells were resuspended in 100 μl of electroporation buffer containing 2 mM HEPES (pH 7.2), 15 mM K2HPO4/KH2PO4, 250 mM manitol, and 1 mM MgCl2 at a final pH of 7.2. DNA (3 μg) were added to the cell suspension and electroporation was conducted in a 0.1 cm cuvette (BioRad) with a Gene Pulser II apparatus and Gene Pulser II RF module (BioRad) at 140 volts, 15 times 1.5 ms burst duration and 1.5 s intervals. Cells were seeded in a 10-cm dish 5 min after electroporation. Cells were washed 16 h after transfection and the experiment was performed either 24 or 48 h later.
Microscopy analysis and quantification of mitochondrial Bid
After treatments, cells were fixed for 20 min with 3% paraformaldehyde at room temperature and analyzed using a Leica TCS NT confocal laser-scanning microscope (Leica Microsystems, Germany). To quantify cells with a punctate green fluorescent pattern (representing mitochondrial localization), at least 100 Bid-GFP expressing cells were counted in different fields. The percentage of punctate cells represents the percentage of cells expressing Bid-GFP at the mitochondria relative to total number of cells expressing Bid-GFP. Mitochondria were stained with 150 mM Mitotracker Red (Molecular Probes) and nuclei were stained with 1 mM TOPRO-3 (Molecular Probes).
Recombinant tBid
The cDNA coding for full-length human Bid cloned into pGEX4T1 was donated by Dr. Donald D Newmeyer (San Diego, CA, USA). The plasmid was transformed into Escherichia coli BL21 and recombinant tBid was produced as described.48
Cell permeabilization experiments
Cells (6 × 105) per well were seeded in six-well plates. The next day cells were washed twice with washing buffer containing 120 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 0.2 mM MgCl2, 0.1 mM EGTA and 20 mM HEPES-NaOH pH 7.4, and then incubated 10 min in permeabilization buffer containing 120 mM KCl, 10 mM NaCl, 1 mM KH2PO4, 20 mM HEPES-Tris pH 7.2, a protease inhibitor cocktail (Roche Diagnostic, GmbH, Mannheim, Germany), 100 μg/ml digitonin (Sigma), 2 mM succinate, 2 mM ATP, 5 mM phosphocreatine (Sigma) and 5 U/ml creatine phosphokinase (Sigma) at 37°C in the presence or absence of 500 ng of recombinant tBid. After incubation, both cells and medium were collected and cyt c content was determined by SDS-PAGE and Western blotting with cyt c antibody.
Abbreviations
- MAPK:
-
mitogen-activated protein kinase
- PKC:
-
protein kinase C
- PDBu:
-
phorbol-12,13-dibutyrate
- TRAIL:
-
tumor necrosis factor-related apoptosis-inducing ligand
- TNF:
-
tumor necrosis factor
- GFP:
-
green fluorescent protein
- BH3:
-
Bcl-2 homology domain 3
- DISC:
-
death-inducing signaling complex
- FADD:
-
Fas-associated death domain
- Cyt c:
-
cytochrome c.
References
Ashkenazi A, Dixit VM (1998) Death receptors: signaling and modulation. Science 281: 1305–1308.
Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hebert A, DeForge L, Koumenis IL, Lewis D, Harris L, Bussiere J, Koeppen H, Shahrokh Z, Schwall RH (1999) Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 104: 155–162.
Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, Krammer PH, Walczak H (2000) FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12: 599–609.
Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17: 1675–1687.
Luo X, Budihardjo I, Zou H, Slaughter C, Wang X (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94: 481–490.
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292: 727–730.
Chai J, Du C, Wu JW, Kyin S, Wang X, Shi Y (2000) Structural and biochemical basis of apoptotic activation by Smac/DIABLO. Nature 406: 855–862.
Zou H, Li Y, Liu X, Wang X (1999) An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274: 11549–11556.
Rodriguez-Tarduchy G, Lopez-Rivas A (1989) Phorbol esters inhibit apoptosis in IL-2-dependent T lymphocytes. Biochem. Biophys. Res. Commun. 164: 1069–1075.
Illera VA, Perandones CE, Stunz LL, Mower Jr DA, Ashman RF (1993) Apoptosis in splenic B lymphocytes. Regulation by protein kinase C and IL-4. J. Immunol. 151: 2965–2973.
Scaffidi C, Schmitz I, Zha J, Korsmeyer SJ, Krammer PH, Peter ME (1999) Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J. Biol. Chem. 274: 22532–22538.
Ruiz-Ruiz C, Robledo G, Font J, Izquierdo M, Lopez-Rivas A (1999) Protein kinase C inhibits CD95 (Fas/APO-1)-mediated apoptosis by at least two different mechanisms in Jurkat T cells. J. Immunol. 163: 4737–4746.
Whitehurst CE, Boulton TG, Cobb MH, Geppert TD (1992) Extracellular signal-regulated kinases in T cells. Anti-CD3 and 4 beta-phorbol 12-myristate 13-acetate-induced phosphorylation and activation. J. Immunol. 148: 3230–3237.
Meng XW, Heldebrant MP, Kaufmann SH (2002) Phorbol 12-myristate 13-acetate inhibits death receptor-mediated apoptosis in Jurkat cells by disrupting recruitment of Fas-associated polypeptide with death domain. J. Biol. Chem. 277: 3776–3783.
Harper N, Hughes MA, Farrow SN, Cohen GM, MacFarlane M (2003) Protein kinase C modulates tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by targeting the apical events of death receptor signaling. J. Biol. Chem. 278: 44338–44347.
Soderstrom TS, Poukkula M, Holmstrom TH, Heiskanen KM, Eriksson JE (2002) Mitogen-activated protein kinase/extracellular signal-regulated kinase signaling in activated T cells abrogates TRAIL-induced apoptosis upstream of the mitochondrial amplification loop and caspase-8. J. Immunol. 169: 2851–2860.
Zhang XD, Borrow JM, Zhang XY, Nguyen T, Hersey P (2003) Activation of ERK1/2 protects melanoma cells from TRAIL-induced apoptosis by inhibiting Smac/DIABLO release from mitochondria. Oncogene 22: 2869–2881.
Sarker M, Ruiz-Ruiz C, Robledo G, Lopez-Rivas A (2002) Stimulation of the mitogen-activated protein kinase pathway antagonizes TRAIL-induced apoptosis downstream of BID cleavage in human breast cancer MCF-7 cells. Oncogene 21: 4323–4327.
Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME (1999) Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286: 1358–1362.
Janicke RU, Sprengart ML, Wati MR, Porter AG (1998) Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 273: 9357–9360.
Ruiz-Ruiz C, Munoz-Pinedo C, Lopez-Rivas A (2000) Interferon-gamma treatment elevates caspase-8 expression and sensitizes human breast tumor cells to a death receptor-induced mitochondria-operated apoptotic program. Cancer Res. 60: 5673–5680.
Ruiz de Almodovar C, Ruiz-Ruiz C, Munoz-Pinedo C, Robledo G, Lopez-Rivas A (2001) The differential sensitivity of Bc1-2-overexpressing human breast tumor cells to TRAIL or doxorubicin-induced apoptosis is dependent on Bc1-2 protein levels. Oncogene 20: 7128–7133.
Werlen G, Jacinto E, Xia Y, Karin M (1998) Calcineurin preferentially synergizes with PKC-theta to activate JNK and IL-2 promoter in T lymphocytes. EMBO J. 17: 3101–3111.
Kelekar A, Thompson CB (1998) Bcl-2-family proteins: the role of the BH3 domain in apoptosis. Trends Cell Biol. 8: 324–330.
Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM (1997) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277: 815–818.
Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI, Goddard AD, Godowski P, Ashkenazi A (1997) Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 277: 818–821.
Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ (1998) Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J. Immunol. 161: 2833–2840.
Sheikh MS, Burns TF, Huang Y, Wu GS, Amundson S, Brooks KS, Fornace AJ, Jr, el-Deiry WS (1998) p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 58: 1593–1598.
Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S (1999) Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res. 59: 734–741.
Sivaraman VS, Wang H, Nuovo GJ, Malbon CC (1997) Hyperexpression of mitogen-activated protein kinase in human breast cancer. J. Clin. Invest. 99: 1478–1483.
Hu Y, Dragowska WH, Wallis A, Duronio V, Mayer L (2001) Cytotoxicity induced by manipulation of signal transduction pathways is associated with down-regulation of Bcl-2 but not Mcl-1 in MCF-7 human breast cancer. Breast Cancer Res. Treat 70: 11–20.
Yeh JH, Hsu SC, Han SH, Lai MZ (1998) Mitogen-activated protein kinase kinase antagonized fas-associated death domain protein-mediated apoptosis by induced FLICE-inhibitory protein expression. J. Exp. Med. 188: 1795–1802.
Erhardt P, Schremser EJ, Cooper GM (1999) B-Raf inhibits programmed cell death downstream of cytochrome c release from mitochondria by activating the MEK/Erk pathway. Mol. Cell Biol. 19: 5308–5315.
Shonai T, Adachi M, Sakata K, Takekawa M, Endo T, Imai K, Hareyama M (2002) MEK/ERK pathway protects ionizing radiation-induced loss of mitochondrial membrane potential and cell death in lymphocytic leukemia cells. Cell Death Differ. 9: 963–971.
Sarker M, Ruiz-Ruiz C, Lopez-Rivas A (2001) Activation of protein kinase C inhibits TRAIL-induced caspases activation, mitochondrial events and apoptosis in a human leukemic T cell line. Cell Death Differ. 8: 172–181.
Marsden VS, Strasser A (2003) Control of apoptosis in the immune system: Bcl-2, BH3-only proteins and more. Annu. Rev. Immunol. 21: 71–105.
Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD (2005) BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17: 525–535.
Boucher MJ, Morisset J, Vachon PH, Reed JC, Laine J, Rivard N (2000) MEK/ERK signaling pathway regulates the expression of Bcl-2, Bcl-X(L), and Mcl-1 and promotes survival of human pancreatic cancer cells. J. Cell Biochem. 79: 355–369.
Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ (2000) Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290: 1761–1765.
Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B, Martinou JC (2001) Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8: 601–611.
Breitschopf K, Zeiher AM, Dimmeler S (2000) Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J. Biol. Chem. 275: 21648–21652.
Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ (2003) Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J. Biol. Chem. 278: 18811–18816.
Majewski N, Nogueira V, Robey RB, Hay N (2004) Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol. Cell. Biol. 24: 730–740.
Pastorino JG, Shulga N, Hoek JB (2002) Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 277: 7610–7618.
Harper N, Farrow SN, Kaptein A, Cohen GM, MacFarlane M (2001) Modulation of tumor necrosis factor apoptosis-inducing ligand- induced NF-kappa B activation by inhibition of apical caspases. J. Biol. Chem. 276: 34743–34752.
Brunet A, Pages G, Pouyssegur J (1994) Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene 9: 3379–3387.
Agami R, Bernards R (2000) Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102: 55–66.
von Ahsen O, Renken C, Perkins G, Kluck RM, Bossy-Wetzel E, Newmeyer DD (2000) Preservation of mitochondrial structure and function after Bid- or Bax-mediated cytochrome c release. J. Cell Biol. 150: 1027–1036.
Acknowledgements
This work was supported by grants from the Ministerio de Educación y Ciencia (SAF2003-00402) and Association for International Cancer Research (AICR-03-031) to AL-R GO-F was supported by a fellowship from the Ministerio de Educación y Ciencia (MEC) and AICR contract.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by GM Cohen
Rights and permissions
About this article
Cite this article
Ortiz-Ferrón, G., Tait, S., Robledo, G. et al. The mitogen-activated protein kinase pathway can inhibit TRAIL-induced apoptosis by prohibiting association of truncated Bid with mitochondria. Cell Death Differ 13, 1857–1865 (2006). https://doi.org/10.1038/sj.cdd.4401875
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401875
Keywords
This article is cited by
-
P38/MAPK contributes to endothelial barrier dysfunction via MAP4 phosphorylation-dependent microtubule disassembly in inflammation-induced acute lung injury
Scientific Reports (2015)
-
Modulation of apoptosis-related cell signalling pathways by curcumin as a strategy to inhibit tumor progression
Molecular Biology Reports (2014)
-
Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis
Cell Death & Differentiation (2013)
-
Sensitization to TRAIL-induced apoptosis in human neuroblastoma SK-N-AS cells by NF-κB inhibitors is dependent on reactive oxygen species (ROS)
Journal of Neuro-Oncology (2011)
-
A mitochondrial block and expression of XIAP lead to resistance to TRAIL-induced apoptosis during progression to metastasis of a colon carcinoma
Oncogene (2008)