Abstract
Recent studies strongly suggest an active involvement of the c-Jun N-terminal kinase (JNK) signaling pathway in tumor necrosis factor (TNF)-induced apoptosis. The direct evidence for the role of JNK and its isoforms has been missing and the mechanism of how JNK actually could facilitate this process has remained unclear. In this study, we show that Jnk2−/− primary mouse embryonic fibroblasts (pMEFs) exhibit resistance towards TNF-induced apoptosis as compared to corresponding wild-type and Jnk1−/− pMEFs. JNK2-deficient pMEFs could be resensitized to TNF via retroviral transduction of any of the four different JNK2 splicing variants. Jnk2−/− pMEFs displayed deficient and delayed effector caspase activation as well as impaired cytosolic cystein cathepsin activity: processes that both were needed for efficient TNF-induced apoptosis in pMEFs. Our work demonstrates that JNK has a central role in the promotion of TNF-induced apoptosis in pMEFs, and that the JNK2 isoform can regulate both mitochondrial and lysosomal death pathways in these cells.
Similar content being viewed by others
Introduction
Exposure of cells to tumor necrosis factor (TNF) can induce both anti- and proapoptotic signaling. Binding of TNF to its receptor complex initiates proapoptotic pathways via the Fas-associated death domain protein(FADD). TNF can concomitantly induce a cellular survival pathway through the TNF receptor complex associating protein TRAF2. This is based on the ability of TRAF2 to mediate signal-specific activation of the transcription factor nuclear factor kappaB (NF−κB), which upon activation initiates the expression of genes that promote cell survival.1,2
TNF exerts its activity via two distinct TNF receptors (TNFR): TNFR1 and TNFR2, which both belong to the TNFR superfamily together with Fas/Apo-1. Both TNFR1 and Fas can transduce apoptotic stimuli via caspase-8 recruitment to the FADD. Upon activation, caspase-8 catalyses the subsequent cleavage-mediated activation and amplification of the effector caspase cascade.3,4 In contrast to Fas that can directly associate with FADD, TNFR1-mediated apoptosis requires an adaptor protein TRADD for the recruitment of FADD.5 While death receptors can directly activate caspases (extrinsic pathway), most other apoptotic stimuli is mediated through the mitochondrial release of cytochrome c (intrinsic pathway; mitochondrial death pathway). In TNFR1/Fas-mediated death signaling, these two pathways can converge via Bcl-2 family member Bid. Bid can be activated via cleavage by caspase-8. Truncated Bid translocates to mitochondria where it induces cytochrome c release into the cytosol, leading to effector caspase activation.6,7
While both the initiation and execution of apoptosis or programmed cell death (PCD) have been previously shown to solely rely on the activation of caspases, recent reports have documented active participitation and even execution of PCD by another family of cysteine proteases, cathepsins.8 Thus, conditions like oxidative stress, growth factor starvation and Fas activation have been shown to induce apoptosis via lysosomal leakage.9 Furthermore, TNF can induce programmed release of active lysosomal proteases including cathepsin B to cytosol.10,11 Hepatocytes isolated from mice harboring genetically inactivated cathepsin B display markedly diminished cytochrome c release, caspase activation and apoptosis upon TNF treatment, suggesting that cathepsin B contributes to the mitochondrial death pathway.10 Furthermore, treatment of the mouse fibrosarcoma cell line WEHI-S with TNF induces increase in the cytosolic cathepsin B activity. The pharmacological inhibition of cathepsin B activity can fully rescue WEHI-S cells from TNF-induced apoptosis, demonstrating that the release of cathepsin B to the cytosol can even act as a dominant executioner of apoptosis.11 It is currently unclear what makes cathepsins to ‘escape’ from lysosomes and what are their cytosolic targets that contribute to apoptotic cell death.9,10 It has been recently suggested that lysosomal proteases, once released to cytosol, could for example be responsible for Bid cleavage.12 Lysosomal proteases would then reside upstream of effector caspases in the apoptotic pathway, and form an additional branch in the apoptotic death pathway, the lysosomal death pathway.
JNK activation, just like TNF signaling, is associated with both apoptosis and survival. Studies utilizing ectopically expressed dominant-negative and dominant-active JNK signaling pathway members as well as studies utilizing antisense technology support JNK involvement in both processes.13,14,15,16,17,18 It is puzzling how JNK can transmit such opposite signals. TNF signaling is proapoptotic only upon simultaneous inhibition of NF-κB-mediated survival pathway, a process, which will subsequently induce prolonged JNK activation. Thus, the length of the JNK activation is believed to be the major determinant of cell fate in respect to JNK signaling in these model systems.17,18,19
JNK family consists of three different isoforms: JNK1, JNK2 and JNK3, which are products of three individual genes. The expression of JNK1 and JNK2 is ubiquitous, while the expression of JNK3 is neural specific.20,21,22 Jnk1 and Jnk2 genes each give rise to four and Jnk3 to two splice variants. One of the splicing targets is the substrate association site and others produce either amino (N)- or carboxy (C)-terminally extended JNK proteins.23 The functional difference between the long and short forms is unknown. Selective expression of splicing variants could account for the differences in biological responses, especially since splicing can account for substrate specificity.21 Studies with genetically inactivated JNKs have demonstrated that none of the three JNK isoforms is necessary for normal mouse development. Double knockouts between Jnk1 and Jnk3 as well as between Jnk2 and Jnk3 are fully viable, but Jnk1 and Jnk2 double knockout dies at embryonic day (E) 10.5, suggesting that the biological functions of JNK1 and JNK2 isoforms are at least partially overlapping during embryonic development.24,25 Interestingly, the Jnk1−/−Jnk2−/− double knockout embryos display an open neural tube due to deregulated apoptosis. This includes reduced apoptosis in the hindbrain neuroepithelium at E9.25, followed by a massive regional increase in cellular apoptosis at both forebrain and hindbrain at later time points, demonstrating that JNKs can transmit both apoptotic and survival signals in vivo.24,25 Adult mice harboring individual Jnk gene disruptions display differences in various biological responses including kainic acid-induced hippocampal exitoxicity, development of obesity, type II diabetes and experimental skin cancer,26,27,28,29 which suggests that individual JNK isoforms have different biological functions in an adult organism.
Studies utilizing Jnk1−/− and Jnk2−/− cell lines largely support the proapoptotic role of JNK. Thus, thymocytes derived from mice with disrupted Jnk genes are partially resistant to CD3-induced apoptosis, and for example JNK2 has been reported to be required for apoptosis of immature thymocytes induced by anti-CD3 antibody.30,31,32 Furthermore, MEFs expressing neither JNK1 nor JNK2 display suppressed apoptosis after ultraviolet light (UV) irradiation or treatment with the genotoxic agent methylmethanesulfonate (MMS).33 UV—light- and MMS-induced apoptosis is also impaired in pMEFs isolated from mice lacking the expression of JNK-activating kinases MKK4 and MKK7.34 Interestingly, however, Fas-mediated apoptosis in both thymocytes and pMEFs has been reported to be independent of JNK.31,33
What is the role of JNK in TNFR1-mediated apoptosis? Recent studies utilizing overexpression of dominant-negative JNK upstream kinases JNKK1 (MKK4/SEK1) or JNKK2 (MKK7) have indirectly demonstrated a proapoptotic role for JNK in TNF-induced death of mouse and human fibroblasts.17,18,35 Furthermore, genetic inactivation of one of the JNKK upstream kinases, Ask1, leads to partial impairment of TNF-induced apoptosis as well as shortens the period of TNF-induced JNK activation.19 Thus far, the possible direct involvement of JNK in TNF-induced apoptosis as well as the mechanism of how JNK could facilitate it has remained unclear. These two important aspects have been addressed in this study.
Utilizing cells with genetically inactivated JNK1 and JNK2 isoforms, we provide direct evidence that JNK2 mediates TNF-induced apoptosis in pMEFs. In this study, we used mice that are on C57BL/6 background in order to obtain TNF-sensitive pMEF populations in which the effect of the genetic background could be excluded. We show that Jnk2−/− pMEFs isolated from three different mouse embryos at E14 were clearly more resistant to TNF treatment than two corresponding individual Jnk1−/− and two wild-type (wt) pMEF lines. Importantly, Jnk2−/− cells could be resensitized to TNF upon retroviral reintroduction of any of the four known JNK2 splice variants. Concomitant with decreased TNF sensitivity, Jnk2−/− pMEFs showed impaired effector caspase activation and deficiency in the release of the B/L-like cytosolic cysteine cathepsins to the cytosol compared to wt pMEFs upon TNF treatment. This deficiency in proapoptotic protease activities correlated with both impaired cytochrome c release and Bid cleavage. With specific protease inhibitors, we demonstrated that inhibition of neither caspase nor cytosolic cysteine cathepsin activity alone was sufficient to rescue cells from TNF-induced death. Full protection from TNF was achieved only by simultaneous inhibition of both protease pathways.
This study provides first direct evidence for the proapoptotic role of JNK in TNF-induced apoptosis of pMEFs. The main conclusions of this study are that both activation of caspases (mitochondrial death pathway) and appearance of enzymatically active lysosomal cathepsins into the cytosol (lysosomal death pathway) are responsible for the TNF-induced apoptosis of pMEFs, and that JNK2 isoform of JNK has a central role in the activation and regulation of both pathways.
Results
JNK2 is the main JNK isoform involved in TNF-induced cell death
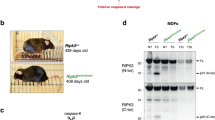
In order to determine the role of JNK1 and JNK2 in cellular signaling, targeted gene disruptions were made of Jnk1 and Jnk2.31,32 The resulting mice were backcrossed into C57BL/6 laboratory mouse strain to obtain TNF-sensitive pMEFs and to avoid the possible effect of the genetic background on TNF sensitivity. Since TNF induces a strong protective signaling via NF-κB activation and consequent gene expression, which in case of fibroblasts will overcome the apoptotic signaling, pMEFs were cotreated with cycloheximide (CHX) to block the protein synthesis in all experiments. Passage 5 pMEFs isolated from two independent wt E14 embryos, two Jnk1 and three Jnk2 knockout embryos were tested for their sensitivity to TNF-induced apoptosis (Figure 1a). All wt and Jnk1 knockout fibroblasts showed clear sensitivity towards 24 h treatment with 0.2 ng/ml TNF and 10 μM CHX as measured by MTT survival assay. All the three Jnk2−/− pMEF cell lines tested in parallel were more resistant towards TNF than the corresponding Jnk1−/− and wt pMEFs. Statistical analysis showed that the difference in the viability between wt and Jnk2−/− pMEFs upon TNF treatment was highly significant (P-value <10−5). 10 μM CHX, which was used throughout the study, blocked protein synthesis efficiently (about 98% block; data not shown) and had only marginal effect on the survival of pMEFs (approximately 95–98% of cells survived after 24 h CHX treatment; data not shown). Nuclear staining and counting of the apoptotic nuclei revealed that the wt cells also exhibited more frequently apoptosis-like nuclear morphologies than the Jnk2−/− cells upon TNF/CHX treatment (Figure 1b). Results of the nuclear staining experiment were consistent with all of our data pointing out that Jnk2−/− cells exhibit deficient and delayed apoptotic response to TNF. To find out the most suitable TNF concentration to be used in further analysis, a dose–response study was carried out with various TNF concentrations (Figure 1c). Result of this experiment shows that Jnk2−/− pMEFs were more resistant to TNF than wt pMEFs over a range of concentrations. We chose to use the lowest concentrations tested, which were sufficient to induce apoptosis in wt pMEFs (0.1 or 0.2 ng/ml, depending on TNF batch used), to be able to focus on the most specific and relevant functions of TNF needed for cell death promotion. The time–response analysis showed that whereas the viability of the wt pMEFs continuously declined during the entire time course measured (from 12 to 24 h), the viability of the Jnk2−/− pMEFs was unaffected from 12 to 20 h after the treatment and was then only moderately reduced after 20 h (Figure 1d). To find out if the observed Jnk2−/− phenotype was due to a specific effect of the absence of JNK2, we subcloned N-terminally HA-tagged JNK2α1, JNK2α2, JNK2β1 and JNK2β2 splicing variant cDNAs into a retroviral pBabe puro vector (pBp) and infected the Jnk2−/− pMEFs either with one of the each different JNK2-expressing viruses or with a pBp control virus. The viability assay showed that the Jnk2−/− pMEFs transduced with any of the different JNK2 splice variants were indeed resensitized to TNF in comparison to the pBp vector control-infected cells (Figure 1e, top panel). Retroviral infection of Jnk2−/− pMEFs increased their resistance to TNF in general, and thus a higher concentration of TNF (10 ng/ml) was used to induce cell death. The expression and activation of all four HA-tagged JNK2 splice variants were verified with immunocomplex kinase assay of the infected pMEFs utilizing the HA-tag (Figure 1e, lower panel). The significance of the sensitization was tested by statistical analysis which showed that the difference of the viability in Jnk2−/− cells infected with JNK2β2 in comparison to the corresponding pBp-infected cells was significant (P-value <10−2; Figure 1f). Data for this analysis was collected from four independent infections of pBp and JNK2β2 into Jnk2−/− pMEFs (each done in triplicate or quadruplicate). The sensitization obtained with transduction of JNK2β2 into Jnk2−/− pMEFs, however, not fully reach the sensitivity of the pBp vector-infected wt pMEFs (Figure 1f). This could be partially due to the fact that the wt cells do express all the different JNK2 splice variants (JT, unpublished observation), and in our experiments we reintroduced each one of them separately. It is also possible that the deprivation of MEFs of an important regulatory protein like JNK2 forces them to compensate this loss in order to survive. Thus, some of the compensation mechanisms could include irreversible alterations in various, as yet uncharacterized cellular functions.
JNK2 knockout pMEFs are resistant to TNF-induced cell death, but can be partially resensitized by retroviral reintroduction of JNK2. (a) Jnk2−/− pMEFs are more resistant to TNF than the corresponding wt cells. MTT viability assays of seven independently isolated passage 5 pMEF cell lines (two wt, two Jnk1−/− and three Jnk2−/−), which were treated with mouse TNF (0,2 ng/ml) and CHX(10 μM) for 24 h. (b) Wt pMEFs exhibit more apoptosis-like nuclear morphologies than Jnk2−/− cells after TNF treatment. Both Jnk2−/− and wt cells were treated with either 0.2 ng/ml of TNF and 10 μM CHX or CHX alone. Cells were stained with Hoechst-33342 and visualized with immunofluoresence microscopy. Percent of condensed apoptotic nuclei was calculated from three parallel experiments. Experiment was done in triplicate. (c) Dose–response of the MTT viability assays of wt and Jnk2−/− pMEFs treated for 24 h with various concentrations of TNF. (d) Time course of the MTT viability assays of wt and Jnk2−/− pMEFs treated with TNF (0,1 ng/ml) and CHX(10 μM). (e) Retroviral introduction of four different JNK2 splice variants into Jnk2−/− cells. Top panel: MTT viability assay of TNF/CHX (10 ng/ml and 10 μM, respectively) treated Jnk2−/− pMEFs infected with HA-tagged JNK2α1, JNK2α2, JNK2β1 or JNK2β2 cDNA viruses, or with pBp vector control virus. Passage 4 JNK2−/− pMEFs were infected with any of the four HA-tagged JNK2 cDNA viruses or with vector control virus and subjected to a period of selection (2.5 μg/ml puromycin). Retroviral JNK2s are expressed and activated by TNF in pMEFs. Jnk2−/− pMEFs were infected with either vector control virus or one of the four HA-tagged JNK2 splice variant cDNA viruses (pBp, JNK2α1, JNK2α2, JNK2β1, JNK2β2) and cell lysates were prepared. Lysates were subjected to immunocomplex kinase assay using the anti-HA-antibody. (f) MTT viability assay comparing the JNK2−/− cells infected with pBp vector control to the same cells infected with J2β2 and to the corresponding wt cells infected with pBp vector control. Infections and selection were performed as described in Figure 1e. All MTT viability assays are representative of three independent experiments done in triplicates
Jnk2−/− pMEFs exhibit impaired caspase and cytosolic cathepsin activity as well as deficient cytochrome c release upon TNF stimulation
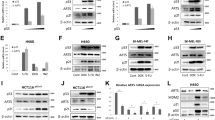
Knowing that TNF activates the caspase pathway, we measured the effector caspase activation in wt, Jnk1−/− and Jnk2−/− pMEFs at various time points after TNF/CHX treatment. To evaluate the caspase-3 like activity upon TNF/CHX treatment, we used a fluorogenic assay measuring the cleavage of Ac-DEVD-AFC fluorogenic probe. The results showed that whereas wt pMEFs displayed a clear rise in DEVDase activity from 4 to 12 h after the TNF treatment, Jnk2−/− pMEFs displayed delayed and decreased effector caspase activation (Figure 2a). In concordance with the impaired effector caspase activation, we also detected deficiency and delay in the cytochrome c release from mitochondria in Jnk2−/− pMEFs in immunostaining experiments (Figure 2b). In the immunofluoresence images, cytochrome c is stained green and it exhibits characteristic granular staining around the nucleus, which is stained red (ethidium bromide). The white arrows on cytochrome c stainings point to the released cytochrome c that results in more diffuse staining. The white arrows on nuclear staining point to condensed, apoptotic nuclei. The images of wt pMEFs after 8 h treatment with 0.2 ng/ml of TNF in presence of CHX show that these cells have clearly increased cytochrome c release and exhibit more condensed nuclei (Figure 2b, image X and image XI) as compared to Jnk2−/− pMEFs (Figure 2b, image IV and image V). No difference was observed between wt and Jnk2−/− pMEFs with the control samples (Figure 2b; compare image I to VII and image II to VIII).
Jnk2−/− pMEFs display deficiency in TNF-induced caspase and cytosolic cathepsin activity, and exhibit deficient cytochrome c release. (a) Jnk2−/− pMEFs exhibit impaired caspase-3 like activity. Wt and Jnk2−/− passage 5 pMEFs were treated with TNF (0.2 ng/ml) plus CHX (10 μM) for various times as indicated. The cell extracts were prepared and caspase-3-like activity was measured using the Ac-DEVD-AFC fluorogenic probe. The figure shows mean and S.D. from three independent experiments (b) Jnk2−/− pMEFs are deficient in cytochrome c release. Jnk2−/− pMEFs (images I–-VI) and wt pMEFs (images VII–XII) were treated for 8 h with CHX alone (images I–III; double staining of Jnk2−/− cells and images VII–IX; double staining of wt pMEFS) or TNF (0.2 ng/ml) and CHX (images IV–VI; double staining of Jnk2−/− cells and images X–XII; double staining of wt cells). Cells were stained to detect cytochrome c (green) and DNA (red). Arrowheads on green show released cytochrome c and arrowheads on red point the condensed apoptotic nuclei. (c) Jnk2−/− pMEFs are deficient in cytosolic cathepsin B/L activity. Wt and Jnk2−/− pMEFs were treated with TNF (0.2 ng/ml) and CHX (10 μM) for various times as indicated. Cytosolic fractions were extracted using a mild extraction buffer containing 15 μg/ml digitonin, and total extracts were prepared using an extraction buffer containing 200 μg/ml digitonin. Cytosolic cysteine cathepsin activities were measured in the cytosolic fractions, and total cellular cytosolic cathepsin activities were measured in total cell extracts using the zFR-AFC fluorogenic probe. Lactate dehydrogenase (LDH) activities were measured both in cytosolic fractions and total extracts and used as internal control for extraction. The figure shows activity values and S.D. from three independent experiments. (d) Jnk2−/− pMEFs are deficient in TNF-induced lysosomal cathepsin B release. Wt and Jnk2−/− pMEFs were treated with CHX and various concentrations of TNF for 8 h. Cytosolic proteins were extracted and cathepsin B present in cytosolic and membrane fractions were evaluated by Western blotting. Alpha-tubulin was used as loading control
A group of lysosomal proteases, cathepsins, which are released from lysosomes to the cytosol during TNF-induced apoptosis, have recently been shown to be responsible for the execution of TNF-induced apoptosis in the murine fibrosarcoma cell line WEHI-S.11 To test if the lysosomal death pathway can also be activated and utilized in pMEFs upon TNF stimulation and whether genetic inactivation of JNK2 has any effect on it, we extracted the cytosolic and total protein fractions of pMEFs and measured the cysteine cathepsin activity (mainly cathepsins B and L) using the fluorogenic probe z-FR-AFC. TNF/CHX treatment was clearly capable of inducing cysteine cathepsin release from lysosomes to cytosol in wt pMEFs as evaluated by appearance of the lysosomal cysteine cathepsin activity in the cytosol (Figure 2c). Furthermore, the cytosolic cysteine cathepsin activity of the Jnk2−/− pMEFs was clearly lower than that of the wt pMEFs throughout the whole experiment (4–16 h of TNF treatment; Figure 2c). The total cysteine cathepsin activity of the wt and Jnk2−/− pMEFs was similar, showing that the lower relative cytosolic cysteine cathepsin activity measured for Jnk2−/− was not due to higher total activity (data not shown). Next, we analyzed the leakage of cathepsin B from the lysosome to the cytosol by Western blotting. Alpha-tubulin was used as a loading control. The disappearance of the cathepsin B from the membrane fraction and its appearance to the cytosolic fraction correlated with the increasing amount of TNF (Figure 2d). Furthermore, the disappearance of cathepsin B from the membrane fraction as well as appearance in the cytosolic fraction was more pronounced in the wt cells than in the Jnk2−/− cells (Figure 2d), and thus correlated well with the data obtained by cysteine cathepsin activity measurements (Figure 2c).
JNK2 is involved in the activation of both mitochondrial and lysosomal death pathways
Since in Jnk2−/− pMEFs both caspase-3 and cytosolic cysteine cathepsin activities were reduced, we decided to examine how the activation of each of these pathways contributes to TNF-induced apoptosis. Thus, we found out that inhibition of caspase activity by zVAD-fmk or cathepsin activity by zFA resulted only in partial rescue (Figure 3a). The specific cathepsin B inhibitor CA074-Me severely sensitized pMEFs to TNF (data not shown). Pretreatment of pMEFs with caspase-8 inhibitor IETD-CHO also had only marginal effect on cell viability (Figure 3a). However, the TNF-induced cell death could be fully blocked when both caspases and cysteine cathepsins were inhibited by cotreatment of cells with low concentration of zVAD-fmk, (specific inhibition of caspases11,36) and zFA-fmk or alternatively with high concentration of zVAD-fmk alone (efficient inhibition of both caspases and cysteine cathepsins;11,36 Figure 3a). Statistical analysis proved that the rescue obtained with low zVAD-fmk and zFA-fmk cotreatment (P-value <10−3) and high zVAD-fmk (P-value <10−4) was highly significant. Effects obtained with low zVAD-fmk, zFA-fmk and IETD-CHO alone as well as IETD-CHO together with zFA-fmk were nonsignificant. Treatment of cells with either zVAD-fmk or zFA-fmk did not have any effect on the TNF-induced JNK activation as measured with the immunocomplex kinase assay (Figure 3b), demonstrating that the rescuing effect exerted by zVAD-fmk and zFA-fmk was not due to unspecific inactivation of JNK by these inhibitors.
TNF-induced apoptosis involves both caspase and cathepsin protease pathways. (a) TNF-induced death of wt pMEFs can be reverted by inhibiting both caspase and cathepsin proteases. MTT viability assay of wt and Jnk2−/− passage 5 pMEFs treated for 20 h with 0.2 ng of TNF and 10 μg/ml CHX in the presence of the 5 μM (low zVAD), 50 μM (high zVAD) zVAD-fmk, 100 μM zFA-fmk or 50 μM IETD-CHO. The values presented here represent mean of three independent experiments. (b) Caspase and cathepsin inhibitors do not inhibit TNF-inducible JNK activity. JNK immunocomplex kinase assay of wt pMEFs treated with TNF/CHX and with or without 5, 50 μM zVAD-fmk or 100 μM zFA-fmk. Cells were lysed and JNK was immunoprecipitated using an anti-JNK antibody (333; Pharmingen). The activity of precipitated JNK was measured using GST-c-Jun (1–79) as substrate. (c) Jnk2−/− pMEFs are deficient in TNF-induced cytochrome c release. Western blots of the cytosolic extracts isolated from TNF/CHX-treated (8 h) wt and Jnk2−/− pMEFs) with and without of 5 μM zVAD-fmk, 100 μM zFA-fmk, both 5 μM zVAD-fmk and 100 μM zFA-fmk, 50 μM zVAD-fmk or 25 μM CA074-Me. Negative control was treated with CHX alone. Expression of GAPDH was used as a control. (d) Jnk2−/− pMEFs are deficient in Bid cleavage. Wt and Jnk2−/− passage 5 pMEFs were treated with TNF/CHX with and without of 5 μM zVAD-fmk, 100 μM zFA-fmk, both 5 μM zVAD-fmk and 100 μM zFA-fmk, or 25 μM CA074-Me for 8 h. Total protein lysates were analyzed by Western blotting using an anti-Bid primary antibody. Expression of GAPDH was used as control
We also measured effect of protease inhibitors on the TNF/CHX-induced appearance of cytochrome c in the cytosol by Western blotting analysis. Cytosolic fractions were prepared from both wt and Jnk2−/− pMEFs treated for 8 h with 0.2 ng/ml of TNF (and CHX) and with or without the presence of selected protease inhibitors (Figure 3c). In accordance with the immunofluoresence data in Figure 2b, TNF/CHX treatment of Jnk2−/− pMEFs did not result in any clearly detectable release of cytochrome c into the cytosol in contrast to wt cells (Figure 3c). Low concentration of zVAD-fmk could at least partially block cytochrome c release in wt cells. No inhibition of the cytochrome c release was detected with cathepsin inhibitors zFA-fmk and CA074-Me (25 μM, specific cathepsin B inhibitor). On the contrary, complete blockage was observed in cells pretreated with low zVAD-fmk together with zFA-fmk or with high zVAD-fmk.
Additionally, in Jnk2−/− cells, we observed deficiency in the cleavage of the proapoptotic Bcl2 family member Bid, which in wt cells was clearly detectable at 8 h time point (Figure 3d, right panel). It seemed like the TNF-induced Bid cleavage in wt pMEFs could be partially inhibited by pretreatment of cells with a low concentration of caspase inhibitor zVAD-fmk (5 μM, specific inhibition of caspases11,36), as could be judged from the full-length Bid (Figure 3d, left panel; lane 3 in comparison to lanes 1 and 2). However, the p15 cleavage fragment of Bid was also clearly detectable (Figure 3d, left panel; lane 3), suggesting that inhibition of caspases was not sufficient to fully block the Bid cleavage. Preincubation of wt pMEFs with neither the cathepsin inhibitor zFA-fmk nor CA074-Me could prevent the Bid cleavage, in contrast to the cotreatment with low concentration of zVAD-fmk and zFA-fmk that could fully block Bid cleavage (Figure 3d). These data suggest that both caspase and cysteine cathepsin activities are contributing to the TNF-induced Bid cleavage in pMEFs.
JNK2 is the major JNK isoform in pMEFs
To determine the effect of the genetic inactivation of Jnk1 and Jnk2 on the total JNK protein levels, a Western blot of equal amounts of protein extracts from wt, Jnk1−/− and Jnk2−/− cells were prepared. Western blot analysis utilizing pan anti-JNK antibody (666, Pharmingen) showed that the JNK expression level in Jnk2−/− pMEFs was clearly reduced as compared to wt cells and also slightly lower than in Jnk1−/− cells (pending that the affinity of the antibody is the same for JNK1 and JNK2; Figure 4a). Since TNF-inducible JNK activity is a functional parameter and thus more relevant than the absolute JNK protein amount, we next measured the JNK activity in wt, Jnk1−/− and Jnk2−/− pMEFs upon TNF stimulation. To find out if the total reduction in the JNK amount has an effect on the TNF inducibility of JNK, passage 5 pMEFs from wt, Jnk1−/− and Jnk2−/− embryos were treated with TNF and CHX for various times, as indicated. The cell lysates were prepared and tested with in-gel kinase assay using recombinant c-Jun (1–79) as a substrate (Figure 4b, upper panel). All three JNK genotypes tested displayed the same activation kinetics, a peak in the activity after 1 h and a drop to near the basal activity after 4 h (Figure 4b, upper panel). Quantitation of the analysis showed that JNK2 accounted for 57% of the total TNF-inducible JNK activity of the pMEFs as estimated from the 1 h treatment point. Furthermore, it showed that most of the JNK2 isoform activity came from the 55 kDa form and most of the JNK1 activity from the 46 kDa form (Figure 4b, lower panel). Interestingly, reintroduction of either of the two 55 kDa JNK2 splice variants (JNK2α2 and JNK2β2) into Jnk2−/− pMEFs resulted in as good sensitization of these cells to TNF as the corresponding short forms (see Figure 1e), suggesting that the JNK isoform composition in the cells could be more important determinant for the PCD signaling than the total JNK amount itself. The data from the in-gel kinase assay demonstrates that the differences in the total JNK activity of the individual isoforms (Figure 4b) correlated closely with the differences in the protein amounts visualized by the Western blot analysis (Figure 4a).
JNK2 is the major JNK isoform in pMEFs isolated from C57BL/6. (a) Western blot analysis of wt, Jnk1−/−, and Jnk2−/− pMEFs using a JNK antibody (666, Pharmingen). Equal amounts of total cell extracts from passage 5 pMEFs were loaded for each genotype. Expression of Hsc 70 was used as a control. (b) Top panel: in-gel kinase assay of wt, Jnk1−/−, and Jnk2−/− pMEFs. pMEFs were harvested and lysed after treatment with TNF/CHX (0.1 ng/ml mTNF/10 μM CHX) for different time periods. In total, 10 μg of each sample was loaded on a gel containing immobilized recombinant GST-c-Jun (1–79), and JNK activity was assessed as described in Material and Methods. Lower panel: quantitations of in-gel kinase assay. Quantitations were done using the Fuji FLA-3000 phosphor imager and the Fuji Image Gauge software. White part of each bar represents the activity of the light JNK splice variants (46 kDa) and black bars represent the activity of the heavy JNK splice variants (55 kDa)
Both TNF receptor and caspase-8 are functional in Jnk2−/− pMEFs
The NF-κB pathway is shown to function independently of the JNK activation in response to TNF receptor signaling.37,38 Thus, in order to verify that the genetic inactivation of either JNK1 or JNK2 did not impair TNF signaling at the receptor level, we measured the NF-κB transcriptional activity via transient expression of an NF-κB luciferase reporter construct with or without 4 h TNF-treatment (10 ng/ml). The TNF-inducible NF-κB activity was not decreased in pMEFs lacking either JNK1 or JNK2, demonstrating that the survival signaling from the TNF receptor is not affected by genetic inactivation of JNK1 and JNK2 (Figure 5a). To assess the possible effect of JNK2 at the level of the initiator caspase (caspase-8) activation, we treated both wt and Jnk2−/− cells with various concentrations of FasL in the presence of CHX and measured their viability. FasL has been reported to induce death solely through activation of the caspase-8 and caspase-3 pathways.39,40 Our results showed that both wt and Jnk2−/− cells were equally sensitive to FasL (Figure 5b), as has also been shown in an earlier study.33 More importantly, we could largely revert the FasL-induced death of Jnk2−/− cells by pretreatment of cells with classical caspase inhibitors; low concentration of caspase inhibitor zVAD-fmk (Figure 5c) as well as high concentration of zVAD-fmk (Figure 5c), but not with the cysteine cathepsin inhibitor zFA-fmk (Figure 5c). The caspase-8-specific inhibitor IETD-CHO repeatedly also gave almost as good rescue as zVAD. The reason why we only achieved 80% rescue instead of 100% with any of the inhibitor treatment is unknown, but could be for example due to a possibility that FasL induces other proteases that cannot be inhibited with the used caspase and cathepsin inhibitors, like for example other lysosomal cathepsins than cathepsin B/L-like cysteine cathepsins. Indeed, a previous study shows that Fas ligation can induce lysosomal leakage, which could be responsible for the deficient rescue observed.9All in all, this data demonstrate that the activity and function of caspase-8 is not generally affected by JNK2 inactivation and furthermore, that unlike caspases, cysteine cathepsins (mainly cathepsins B/L) do not have such a central role in the FasL-induced death of Jnk2−/− pMEFs.
JNK2 functions downstream of the TNFR and caspase-8 activation. (a) Genetic inactivation of JNK1 or 2 isoforms does not reduce NF-κB inducibility in pMEFs. Wt, Jnk1−/− and Jnk2−/− passage 5 pMEFs were cotransfected with the NF-κB responsive fire-fly luciferase construct (pHII-fluc) and the Renilla luciferase construct pRL-null. pMEFs were treated with mTNF (10 ng/ml) for 4 h and expression of the luciferase was assessed (Promega, Dual Luciferase Assay). Each bar represents quantitation of three independent experiments (b) JNK2 deletion has no influence on the FasL-induced death of pMEFs. MTT viability assay of wt and Jnk2−/− passage 5 pMEFs treated with varying doses of FasL (as indicated) and CHX (1 μM). (c) FasL/CHX-induced death can be reverted by blocking the caspase activation. MTT viability assay of Jnk2−/− passage 5 pMEFs treated for 20 h with 5% Fas ligand (FasL) media and 1 μg/ml CHX in the presence of either a low concentration of the caspase inhibitor zVAD-fmk (5 μM, low zVAD), a high concentration of zVAD-fmk (50 μM, high zVAD), the cathepsin inhibitor zFA-fmk (100 μM, zFA), the caspase-8 inhibitor IETD-CHO (50 μM) or in the presence of both zVAD-fmk (5 μM) and zFA-fmk (100 μM) or IETD-CHO and zFA-fmk. Each assay was done in triplicate
Discussion
Treatment of fibroblasts with TNF alone induces JNK activity, but does not lead to PCD, showing that cell death is not a mandatory consequence of JNK activity. However, when the TNF-induced activation of the antiapoptotic NF-κB pathway is inhibited either by expression of nondegradable IκB mutant or by inhibition of de novo protein synthesis with for example CHX, cells will undergo apoptosis upon TNF treatment. One of the first measurable events of this phenomenon is that JNK activation becomes prolonged, presumably due to the absence of NF-κB target gene products that are involved in JNK inhibition.17,18,19 While the duration and magnitude of total JNK activity seems to correlate well with the cell fate in response to TNF, the molecular mechanisms behind the proapoptotic action of JNK have remained unclear.41
In this study, we demonstrated that Jnk2−/− pMEFs isolated from E14 mouse embryos were clearly more resistant to TNF than corresponding Jnk1−/− or wt cells. Furthermore, we showed that retroviral reintroduction of any of the four different JNK2 splice variants into Jnk2−/− cells could resensitize them to TNF. Results from both survival assays of individual JNK knockout cells and from the retroviral reintroduction of JNK2 into JNK2-null fibroblasts strongly suggest that JNK2 is the most proapoptotic isoform of JNK in pMEFs with respect to TNF signaling. Concomitantly, we showed that the JNK2 isoform is only slightly more abundant than JNK1 in these cells (according to in-gel kinase assay, JNK2 accounts approximately for 57% of TNF-inducible total JNK activity). It is, however, unlikely that the reduced total JNK concentration and the subsequently reduced total activity were the most critical determinants for the TNF resistance of Jnk2−/− pMEFs, since the difference between TNF-inducible total JNK activity in Jnk1−/− and Jnk2−/− pMEFs was only few percents. Furthermore, Jnk2−/− pMEFs were clearly more resistant over a wide range of TNF concentration than corresponding Jnk1−/− cells. Jnk2−/− pMEFs do however eventually die with TNF within longer time period. This is probably at least partially due to the fact that these cells still have JNK1, which might be able to partially compensate for the loss of JNK2.
Surprisingly, in a total opposition to our study and to both earlier studies utilizing MEFs,17,18 a recent paper showed that inactivation of JNK (either JNK1 or JNK2) sensitizes MEFs to TNF.42 We are currently unable to explain this difference. However, it is plausible that differences in the mouse background contribute to TNF sensitivity. According to our experience, early passage C57BL/6 pMEFs are much more sensitive to TNF treatment than C57BL/6-129 mixed background pMEFs, a difference which diminishes upon immortalization (data not shown). Hochedlinger and co-workers used pMEFs isolated from C57BL/6-129 mixed background embryos (K Sabapathy, personal communication). Poor TNF sensitivity of the mixed background strain might explain why they needed to use 5000 times higher concentration of TNF than we to induce death in pMEFs. If the poor TNF sensitivity of the C57BL/6-129 mixed background pMEFs is due to TNF-receptor amount or some other possible differences in TNF signaling pathway is unknown, but ectopic expression of TNFR1 has been used as a general way to overcome it.43
How is JNK involved in TNF-induced apoptosis? Previously, JNK has been shown to participate in the mitochondrial death pathway in the UV-induced cell death.33 Treatment of fibroblasts with TNF also activates both JNK and the mitochondrial death pathway, as shown in this study and in an earlier study involving human primary fibroblasts.35 In agreement with this, we have shown that Jnk2−/− pMEFs display impaired Bid cleavage, mitochondrial cytochrome c release and effector caspase activation, suggesting that JNK2 is upstream of the mitochondrial death pathway in pMEFs (Figure 6). Nevertheless, specific inhibition of caspases had only minor effect on viability, indicating that an alternative death pathway is activated upon TNF induction. Furthermore, the specific inhibition of cysteine cathepsins, a group of lysosomal proteases that can act as executioners in TNF-induced cell death in WEHI-S mouse fibrosarcoma cell line,11 did also only partially rescue pMEFs from TNF-induced death. However, simultaneous inhibition of both caspases and cathepsins was sufficient for full rescue. Consistently, both cytosolic cysteine cathepsin activity as well as caspase activity was clearly impaired in Jnk2−/− cells as compared to the corresponding wt cells. Our data show that in pMEFs either caspase activation or appearance of active lysosomal cathepsins into the cytosol is enough for TNF-induced apoptosis, suggesting that two death pathways exist in these cells and that each of these pathways is able to bypass the other and cause apoptosis. One pathway is the mitochondrial death pathway activated by Bid cleavage and successive cytochrome c release followed by the activation of the effector caspases. The other is the cysteine cathepsin-dependent lysosomal death pathway for which the initiators are unknown, but which also seems to involve Bid cleavage. Our data show that JNK2 resides upstream of these pathways and can control both of them (Figure 6). Interestingly, a previous study demonstrates that TNF-induced cytochrome c release from mitochondria into the cytosol is reduced in catB−/− hepatocytes and thus places cathepsin B upstream of the effector caspase activation and mitochondrial death pathway in these cells.10 Our study shows that both Bid cleavage and cytochrome c release were impaired in Jnk2−/− pMEFs placing JNK2 upstream of the mitochondrial death pathway. Furthermore, our work clearly shows that both TNF-induced effector caspase activation and cytosolic cysteine cathepsin activity were impaired in Jnk2−/− pMEFs, thus placing JNK2 upstream of both death pathways. The fact that we could only obtain full rescue of pMEFs by simultaneous inhibition of both pathways, however, further suggests that these two pathways can function in parallel in respect to JNK signaling in pMEFs.
Schematic model of the role of JNK2 in TNF-induced apoptosis of pMEFs based on this study. The novel data provided emphasize a central role for JNK signaling in mediation of the TNF-induced apoptosis in pMEFs by placing JNK upstream of both lysosomal and mitochondrial death pathways. The most important JNK isoform in this model is JNK2
It seems that TNF-induced PCD involves different death pathways, and that the lack of one of these can be compensated by another pathway. Caspase-8 however seems to be central to all of these pathways, since caspase-8 knockout fibroblasts are reported to be fully resistant to TNF-induced death.44 Interestingly, although Jnk2 knockout pMEFs were fully competent for FasL-induced caspase-8 activation, the TNF-inducible cleavage of the proapoptotic protein Bid, which is considered to be a classical caspase-8 substrate, was deficient in Jnk2−/− cells. Bid has been suggested to undergo post-translational modification prior to its cleavage by caspase-8.45 Our data suggest that JNK could be involved in Bid modification in TNF-induced PCD. Bid itself does not have consensus JNK phosphorylation sites and thus it seems more likely that JNK could for example be involved in the phosphorylation and regulation of an enzyme(s) that modifies Bid in pMEFs. Interestingly, it has also been recently shown that lysosomal lysates contain proteinase activity that can cleave Bid in vitro, and thus it has been suggested that cathepsins or other lysosomal proteases in addition to caspase-8 could be involved in Bid cleavage.12 Indeed, several different cathepsins can cleave Bid into its active form in vitro (B Turk, personal communication). This would also explain the fact that we could not fully block Bid cleavage by inhibiting neither caspases nor cathepsins alone.
This study provides novel data, which emphasize a central proapoptotic role for JNK signaling in the promotion of the TNF-induced apoptosis of pMEFs. The most important JNK isoform in this model is JNK2. Our study contributes to the understanding of how JNK is involved in TNF-induced apoptosis of pMEFs and places JNK and especially JNK2 isoform in a central position, upstream of both mitochondrial and lysosomal death pathways, but downstream of TNF–receptor complex formation (Figure 6).
Materials and Methods
Reagents
DMEM that was used for culturing all the cells was purchased from Life Technologies, fetal calf serum from Sigma and mouse TNF from R&D Systems. FasL43-cells expressing Fas ligand were introduced elsewhere.46,47 Protease inhibitor z-Val-Ala-DL-Asp-CH2F (zVAD-fmk) was purchased from Bachem, z-Phe-Ala-CH2F (zFA) from Enzyme System Product, CA074-Me from Peptides International and acetyl-Asp-Glu-Val-Asp-aldehyde (IETD-CHO) from Biomol. Plasmid pEGFP-C3 was purchased from Clontech and pRL-null from Promega. Mouse monoclonal antibodies anti-JNK (333.8), anti-JNK (666) and anti-cytochrome c antibody were purchased from Pharmingen. Mouse monoclonal anti-GAPDH was purchased from Biogenesis. Goat anti-Bid antibody was purchased from R&D Systems and anti-alpha-tubulin from Sigma. Anti-Hsc-70 antibody was a gift from BA Margulis and the cathepsin B antibody from E Weber.
Construction of expression plasmids
The pBp HA vector was constructed by annealing an HA sense-strand with 5′ BamHI overhang (5′-gatccatggcatacccatacgatgttccagattacgctagcg-3′) and a single-stranded HA antisense with EcoRI 5′ overhang (5′-aattcgctagcgtaatctggaacatcgtatgggtatgccatg-3′). Single-stranded HA sense and antisense were mixed in equal amounts, heated to 75°C and left to anneal for 2 h at 25°C and ligated to pBp vector between BamHI and EcoRI site. Murine JNK2α2 and JNK2β2 cDNAs were amplified with polymerase chain reaction (PCR) using primers 5-gaattcatgagtgacagtaaaagc-3′ and 5′-gaattcatatcaccggcagccttcca-3′, the latter of which was replaced by primer 5′-gaattctagccttgctacttactg-3′ for amplification of the corresponding short forms JNK2α1 and JNK2β1. The primers were designed to include 5′-EcoRI sites. The resulting DNA fragments were purified and subcloned into pCR2.1-TOPO vector using the TOPO TA-cloning kit (Invitrogen), after which the JNK2 inserts were cut out with EcoRI, purified and ligated to EcoRI site in pBp.
Isolation, maintenance and metabolic labeling of embryonic fibroblasts
C57BL/6 mice heterozygous for JNK1 and JNK2 and their corresponding mutated allele30,31 were crossed. Embryos were harvested at E12 or E14, fibrous tissues were collected, trypsinized and cultured in DMEM supplemented with nonessential amino acids (4 μg/ml), gentamycin (10 μg/ml) and 10% fetal calf serum in an atmosphere containing 10% CO2.
Genotyping
The DNA was isolated from the embryonic heads according to standard protocol. Isolated DNA was subjected to genotyping utilizing polymerase chain reaction. Oligos JNK1 forward (5′-tcgacccatgctaagcgcgcc-3′) and JNK1 back (5′-ctacttaataacgggggtggagga-3′) were used to amplify the endogenous JNK1 gene fragment and the JNK1 forward together with J1-lacZ (5′-cgggtgcgggcctcttcgc-3′) to detect the mutated JNK1 allele. Oligos JNK2 forward (5′-cgaagcagcagccctcaggatcc-3′) and JNK2 back (5′-ggttctgacgtcctgggctggac-3′) were used to detect the endogenous JNK2, and the same JNK2 forward oligo was used together with J2-lacZ oligo (5′-gcctccagtacagcgcggctg-3′) to detect the JNK2 inactivation. The PCR to detect JNK1 were run as following: 2 min initial denaturation at 95°C followed by 35 cycles of 15 s denaturation at 95°C, 15 s annealing at 62°C and 30 s extension at 72°C. JNK2 detection was performed using 2 min initial denaturation at 95°C followed by 35 cycles of 15 s denaturation at 95°C, 15 s annealing at 60°C and 45 s extension at 72°C. Both JNK1 and JNK2 PCR reactions included a 5 min final extension at 72°C.
Retroviral gene transfer
The PhoenixEco packaging cells were cultured in DMEM, supplemented with 10% FCS, 100 μg/ml of streptomycin and 100 U/ml penicillin. At 24 h prior to transfection, the cells were plated with the density 2 × 106 cells on a 10 cm plate. At 15–20 min prior to transfection, chloroquine was added to 25 μM final concentration, and the retroviral constructs were transfected together with pEGFP-C3 into the cells using the Ca-phosphate coprecipitation method. After 2 days, the retroviral supernatant was harvested, filtered, and polybrene was added to a final concentration of 8 μg/ml. For infection, fibroblasts were incubated with the retroviral media for 2–3 days, which was then replaced by selection media (fibroblast media supplemented with 2.5 μg/ml of puromycin). Target fibroblasts were kept in selection until cultures were confluent, after which the cells were trypsinized and plated for experiments.
Transient transfections and reporter assays
For reporter assays, fibroblasts were cotransfected with pRL-null (Promega) renilla luciferase internal standard plasmid and NF-κB-responsive pBIIX-Luc firefly luciferase reporter plasmid48 constructs using the lipofectamine plus (Gibco-BRL) transfection system. Transfected cells were either treated or not with TNF 3 h prior to harvesting. The cell lysates were prepared and luciferase activities measured according to the manufacturer's protocol (Dual Luciferase Assay – system; Promega).
Viability assays
For MTT-viability assay, the cells were plated with the density of 10 000 cells/well in 96-well plate 24 h prior to treatment. Cells were treated as indicated. The assay was performed as described previously,49 and all treatment conditions were evaluated as triplicates. For LDH assay, cells were plated as described and 50 μl of extract per sample was transferred to 96-well plates (Nunc) and 50 μl of LDH assay reaction mix was added, but in other aspects performed according to the manufacturer's protocol (Roche), and the color formation was measured on a Versamax microplate reader. For Hoechst –staining, cells were plated 50 000 cells/well in 24-well plate 24 h prior to treatment. Cells were treated as indicated and stained 15 min at 37°C with Hoechst-33342 stain, visualized and counted utilizing inverted fluoresence microscope. All viability assays were performed in triplicate or quadruplicate.
Caspase and cathepsin activity measurements
A total of 30 000 cells/well were plated and cultured in 12-well plates 2 days prior to TNF treatment. Cells were treated as indicated transferred into ice and the media was removed. In total, 300 μl of extraction buffer (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 8 mM DTT, 1 mM pefablock, pH 7.5) with either 15 μg/ml (for cytosolic fraction) or 200 μg/ml (for total protein) of digitonin was added. After 12 min incubation on ice, 250 μl of the supernatant was transferred to a microtiter plate. To measure the cysteine cathepsins and caspase 3/7 activity, 50 μl of extract per sample per assay was transferred to black Costar 96-well plates into either 50 μl of 2 × caspase reaction buffer (20% V/V glycerol, 5 mM DTT, 0.5 mM EDTA, 100 mM HEPES, pH 7.0) containing the Ac-DEVD-AFC (50 μM; Enzyme Systems Products) caspase substrate or 50 μl of 2 × cathepsin reaction buffer (50 mM sodium acetate, 8 mM EDTA, 8 mM DTT, 1 mM pefablock, pH 5.0) with the zFR-AFC (50 μM; Enzyme Systems Products) cathepsin substrate. Plates were prewarmed for 5 min to 30°C and the light emission (max. 489 nm, cut off at 475 nm; excitation at 400 nm) was measured on a SpectraMax Gemini fluorescent reader every 2 min for 30 min total.
Preparation of cell lysates for Western blots and kinase assays
To isolate cytosolic extracts for cytochrome c release detection, 500 000 cells/well were plated and cultured in six-well plates 2 days prior to TNF treatment. Cells were treated as indicated, transferred to ice, the media was removed and cells were washed once in PBS. In total, 500 μl of extraction buffer (250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 8 mM DTT, 1 mM befablock, pH 7.5) with 75 μg/ml digitonin was added to each well. After 15 min incubation on ice, extraction buffer was saved and cells were additionally extracted for 15 min on ice using 500 μl extraction buffer without digitonin. Protein was precipitated in 10% TCA and pellet was centrifuged down at 10 000 g for 10 min. Pellets were resuspended in 30 μl urea Laemmli sample buffer, sonicated and run on 15% acrylamide gel. Lysates for cathepsin B detection were prepared as lysates for cathepsin activity measurements. For Bid and JNK, Western blots as well as kinase assays, the cell extracts were prepared as described previously. Western blotting was performed according to standard conditions.
Kinase assays
The in-gel kinase assays were performed as described previously.21 For in-gel kinase assays, 10 μg of cell extract were run on 10% SDS-PAGE with 0.5 mg/ml of GST-cJun polymerized into the gel. For immunocomplex kinase assays, HA-tagged JNKs precipitated from cell lysates using anti-HA-antibody (CA5) and the endogenous JNK was precipitated using JNK antibody (333.8; Pharmingen). The immunocomplex kinase assays were performed as described previously.50
Immunofluoresence microscopy
For cytochrome c staining, the Jnk2−/− and wt cells were plated (50 000 cells/well) on coverslips 24 h prior to treatment in 24-well plates. Cells were treated as indicated and fixed in 4% formaldehyde in PBS for 20 min at room temperature. Cells were permeabilized in 0.2% Triton-X-100 in PBS for 10 min at room temperature, after which they were blocked with 10% fetal calf serum in PBS for 30 min incubation at room temperature. Next, the cells were treated with mouse cytochrome c antibody (Pharmingen) at 1 : 400 dilutions in 0.1% Triton-X-100, 0.25% bovine serum albumin in PBS for 1 h at room temperature. Cells were washed three times for 10 min in 0.1% Triton-X-100, 0.25% bovine serum albumin in PBS at room temperature after which they were incubated with the donkey anti-mouse secondary antibody (Molecular Probes, Alexa-488-coupled, 1 : 1000) in 0.1% Triton X-100, 0.25% bovine serum albumin in PBS for 30 min at room temperature. Cells were washed again one time for 10 min in 0.1% Triton-X-100, 0.25% bovine serum albumin in PBS and treated with 0.5 mg/ml of DNAse-free RNAse for 10 min at 37°C. Cells were washed again once with 0.1% Triton-X-100, 0.25% bovine serum albumin in PBS as before and the DNA was stained with 5 μg/ml of etidium bromide in PBS for 2–3 min at room temperature and mounted with mounting solution (Molecular Probes, Anti-Fade kit) after 10 min final wash in room temperature with Triton-X-100, 0.25% BSA in PBS. The fluorescence images were captured and processed using Zeiss 510 laser-scanning microscope mounted on Axiovert 100M.
Abbreviations
- Ask1:
-
apoptosis signaling kinase 1
- C:
-
carboxy
- CHX:
-
cycloheximide
- E:
-
embryonic day
- FADD:
-
Fas-associated death domain protein
- N:
-
amino
- NF-κB:
-
nuclear factor kappaB
- JNK:
-
c-Jun N-terminal kinase
- JNKK:
-
JNK kinase
- MMS:
-
methylmethanesulfonate
- pBp:
-
pBabe puro
- PCD:
-
programmed cell death
- pMEF:
-
primary mouse embryonic fibroblasts
- TNF:
-
tumor necrosis factor
- TNFR:
-
TNF receptor
- UV:
-
ultraviolet
- wt:
-
wild type
References
Baud V and Karin M (2001) Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 11: 372–377
Chen G and Goeddel DV (2002) TNF-R1 signaling: a beautiful pathway. Science 296: 1634–1635
Ashkenazi A and Dixit VM (1999) Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11: 255–260
Wallach D (1997) Cell death induction by TNF: a matter of self control. Trends Biochem Sci. 22: 107–109
Hsu H, Shu HB, Pan MG and Goeddel DV (1996) TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84: 299–308
Li H, Zhu H, Xu CJ and Yuan J (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94: 491–501
Luo X, Budihardjo I, Zou H, Slaughter C and Wang X (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94: 481–490
Leist M and Jaattela M (2001) Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell. Biol. 2: 589–598
Brunk U and Svensson I (1999) Oxidative stress, growth factor starvation and Fas activation may all cause apoptosis through lysosomal leak. Redox Rep. 4: 3–11
Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, Kaufmann SH and Gores GJ (2000) Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Invest. 106: 1127–1137
Foghsgaard L, Wissing D, Mauch D, Lademann U, Bastholm L, Boes M, Elling F and Jaattela M (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153: 999–1010
Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, Ellerby LM, Bredesen D, Freeze H, Abrahamson M, Bromme D, Krajewski S, Reed JC, Yin XM, Turk V and Salvesen GS (2001) Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J. Biol. Chem. 276: 3149–3157
Xia Z, Dickens M, Raingeaud J, Davis RJ and Greenberg ME (1995) Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science 270: 1326–1331
Chen YR, Wang X, Templeton D, Davis RJ and Tan TH (1996) The role of c-Jun N-terminal kinase (JNK) in apoptosis induced by ultraviolet C and gamma radiation. Duration of JNK activation may determine cell death and proliferation. J. Biol. Chem. 271: 31929–31936
Verheij M, Bose R, Lin XH, Yao B, Jarvis WD, Grant S, Birrer MJ, Szabo E, Zon LI, Kyriakis JM, Haimovitz-Friedman A, Fuks Z and Kolesnick RN (1996) Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature 380: 75–79
Potapova O, Gorospe M, Dougherty RH, Dean NM, Gaarde WA and Holbrook NJ (2000) Inhibition of c-Jun N-terminal kinase 2 expression suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner. Mol. Cell. Biol. 20: 1713–1722
De Smaele E, Zazzeroni F, Papa S, Nguyen DU, Jin R, Jones J, Cong R and Franzoso G (2001) Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signaling. Nature 414: 308–313
Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M and Lin A (2001) Inhibition of JNK activation through NF-kappaB target genes. Nature 414: 313–317
Tobiume K, Matsuzawa A, Takahashi T, Nishitoh H, Morita K, Takeda K, Minowa O, Miyazono K, Noda T and Ichijo H (2001) ASK1 is required for sustained activations of JNK/p38 MAP kinases and apoptosis. EMBO Rep. 2: 222–228
Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M and Davis RJ (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76: 1025–1037
Kallunki T, Su B, Tsigelny I, Sluss HK, Derijard B, Moore G, Davis R and Karin M (1994) JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 8: 2996–3007
Mohit AA, Martin JH and Miller CA (1995) p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron 14: 67–78
Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B and Davis RJ (1996) Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15: 2760–2770
Kuan CY, Yang DD, Samanta Roy DR, Davis RJ, Rakic P and Flavell RA (1999) The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron 22: 667–676
Sabapathy K, Jochum W, Hochedlinger K, Chang L, Karin M and Wagner EF (1999) Defective neural tube morphogenesis and altered apoptosis in the absence of both JNK1 and JNK2. Mech. Dev. 89: 115–124
Yang DD, Kuan CY, Whitmarsh AJ, Rincon M, Zheng TS, Davis RJ, Rakic P and Flavell RA (1997) Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature 389: 865–870
Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M and Hotamisligil GS (2002) A central role for JNK in obesity and insulin resistance. Nature 420: 333–336
She QB, Chen N, Bode AM, Flavell RA and Dong Z (2002) Deficiency of c-Jun-NH (2)-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 62: 1343–1348
Chen N, Nomura M, She QB, Ma WY, Bode AM, Wang L, Flavell RA and Dong Z (2001) Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 61: 3908–3912
Rincon M, Whitmarsh A, Yang DD, Weiss L, Derijard B, Jayaraj P, Davis RJ and Flavell RA (1998) The JNK pathway regulates the in vivo deletion of immature CD4 (+)CD8(+) thymocytes. J Exp Med. 188: 1817–1830
Sabapathy K, Hu Y, Kallunki T, Schreiber M, David JP, Jochum W, Wagner EF and Karin M (1999) JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 9: 116–125
Sabapathy K, Kallunki T, David JP, Graef I, Karin M and Wagner EF (2001) c-Jun NH2-terminal kinase (JNK)1 and JNK2 have similar and stage dependent roles in regulating T cell apoptosis and proliferation. J. Exp. Med. 193: 317–328
Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA and Davis RJ (2000) Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288: 870–874
Tournier C, Dong C, Turner TK, Jones SN, Flavell RA and Davis RJ (2001) MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 15: 1419–1426
Gabai VL, Mabuchi K, Mosser DD and Sherman MY (2002) Hsp72 and stress kinase c-jun N-terminal kinase regulate the bid-dependent pathway in tumor necrosis factor-induced apoptosis. Mol. Cell. Biol. 22: 3415–3424
Schotte P, Declercq W, Van Huffel S, Vandenabeele P and Beyaert R (1999) Non specific effects of methyl ketone peptide inhibitors of caspases. FEBS Lett. 442: 117–121
Liu ZG, Hsu H, Goeddel DV and Karin M (1996) Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-kappaB activation prevents cell death. Cell 87: 565–576
Natoli G, Costanzo A, Ianni A, Templeton DJ, Woodgett JR, Balsano C and Levrero M (1997) Activation of SAPK/JNK by TNF receptor 1 through a noncytotoxic TRAF2-dependent pathway. Science 275: 200–203
Muzio M, Chinnaiyan AM, Kischkel FC, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME and Dixit VM (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85: 817–827
Ashkenazi A and Dixit VM (1999) Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11: 255–260
Lin A (2003) Activation of the JNK signaling pathway: breaking the brake on apoptosis. BioEssays 25: 17–24
Hochedlinger K, Wagner EF and Sabapathy K (2002) Differential effects of JNK1 and JNK2 on signal specific induction of apoptosis. Oncogene 21: 2441–2445
Yeh WC, Pompa JL, McCurrach ME, Shu HB, Elia AJ, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, El-Deiry WS, Lowe SW, Goeddel DV and Mak TW (1998) FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 279: 1954–1958
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O, Lapidot T, Soffer D, Sobe T, Avraham KB, Goncharov T, Holtmann H, Lonai P and Wallach D (1998) Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9: 267–276
Desagher S, Osen-Sand A, Montessuit S, Magnenat E, Vilbois F, Hochmann A, Journot L, Antonsson B and Martinou JC (2001) Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8: 601–611
Rensing-Ehl A, Frei K, Flury R, Matiba B, Mariani SM, Weller M, Aebischer P, Krammer PH and Fontana A (1995) Local Fas/APO-1 (CD95) ligand-mediated tumor cell killing in vivo. Eur. J. Immunol. 25: 2253–2258
Schneider P, Holler N, Bodmer JL, Hahne M, Frei K, Fontana A and Tschopp J (1998) Conversion of membrane-bound Fas(CD95) ligand to its soluble form is associated with downregulation of its proapoptotic activity and loss of liver toxicity. J. Exp. Med. 187: 1205–1213
Saksela K, Cheng G and Baltimore D (1995) Proline-rich (PxxP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 14: 484–491
Jaattela M, Benedict M, Tewari M, Shayman JA and Dixit VM (1995) Bcl-x and Bcl-2 inhibit TNF and Fas-induced apoptosis and activation of phospholipase A2 in breast carcinoma cells. Oncogene 10: 2297–2305
Holmberg C, Katz S, Lerdrup M, Herdegen T, Jaattela M, Aronheim A and Kallunki T (2002) A novel specific role for I kappa B kinase complex-associated protein in cytosolic stress signaling. J. Biol. Chem. 277: 31918–31928
Acknowledgements
We thank Katsuji Yoshioka and Michael Karin for 4xAP-1 luciferase as well as murine JNK2α2 and JNK2β2 cDNAs. We thank Randall Johnson and Michael Karin as well as UCSD animal facilities for the help with the transfer of the mice to Copenhagen, and furthermore we also thank Ingrid Fossar Larsen and the personnel in the mice facilities of Danish Cancer Society for excellent assistance. This work was supported by grants from Novo Foundation, Danish Cancer Research Foundation, Danish Cancer Society, Research Foundation of Leo Pharmaceuticals as well as Bøje Benzon Stipend of Alfred Benzon Foundation (all to TK), the Henselt Foundation of the University of Kiel (CH and TH), the Deutsche Forschungsgemeinschaft supported SFB 415 (CH and TH), and the Faculty of Health Sciences, University of Copenhagen (MGH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by GM Cohen
Rights and permissions
About this article
Cite this article
Dietrich, N., Thastrup, J., Holmberg, C. et al. JNK2 mediates TNF-induced cell death in mouse embryonic fibroblasts via regulation of both caspase and cathepsin protease pathways. Cell Death Differ 11, 301–313 (2004). https://doi.org/10.1038/sj.cdd.4401353
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401353
Keywords
This article is cited by
-
Clostridioides difficile toxin B alone and with pro-inflammatory cytokines induces apoptosis in enteric glial cells by activating three different signalling pathways mediated by caspases, calpains and cathepsin B
Cellular and Molecular Life Sciences (2022)
-
Identification of a c-Jun N-terminal kinase-2-dependent signal amplification cascade that regulates c-Myc levels in ras transformation
Oncogene (2012)
-
NFκB signaling is important for growth of antiestrogen resistant breast cancer cells
Breast Cancer Research and Treatment (2012)
-
Jnk2
AfCS-Nature Molecule Pages (2010)
-
Regulation of apoptosis-associated lysosomal membrane permeabilization
Apoptosis (2010)