Abstract
Links between genes involved in development, proliferation and apoptosis have been difficult to establish. In the Drosophila wing disc, the vestigial (vg) and the scalloped (sd) gene products dimerize to form a functional transcription factor. Ectopic expression of vg in other imaginal discs induces outgrowth and wing tissue specification. We investigated the role of the VG–SD dimer in proliferation and showed that vg antagonizes the effect of dacapo, the cyclin-cdk inhibitor. Moreover, ectopic vg drives cell cycle progression and in HeLa cultured cells, the VG–SD dimer induces cell proliferation per se. In Drosophila, ectopic vg induces expression of dE2F1 and its targets dRNR2 and string. In addition vg, but not dE2F1, interacts with and induces expression of dihydrofolate reductase (DHFR). Moreover, a decrease in VG or addition of aminopterin, a specific DHFR inhibitor, shift the dorso-ventral boundary cells of the disc to a cell death sensitive state that is correlated with reaper induction and DIAP1 downregulation. This indicates that vg in interaction with dE2F1 and DHFR is a critical player for both cell proliferation and cell survival in the presumptive wing margin area.
Similar content being viewed by others
Introduction
Cell fate specification during development relies largely on positional information. This specification is controlled by cell cycle regulators that allow arrest of proliferation at defined times during development. Conversely, little is known about how developmental cues can act on genes driving the cell cycle. The developing wing of Drosophila melanogaster provides a relevant model to identify possible links between developmental pathways and gene products that allow cell survival and cell cycle progression.
The wing originates during embryogenesis from a primordium which proliferates rapidly and homogeneously until the beginning of the third instar.1 Wing growth is patterned by the establishment of compartments of specialized cells, marked by boundaries. The antero-posterior (A/P) compartments are established early during embryogenesis under the control of the engrailed (en) selector gene that regulates the decapentaplegic (dpp) morphogen. The dorso-ventral (D/V) compartments, established at the second instar, are defined by cells respectively expressing or not apterous (ap).2 This leads to the activation of the Notch receptor by its ligands at the D/V compartment boundary.3
Several studies point to the vestigial (vg) gene as the main target for the patterning systems defined by the two axes of the wing.4,5,6 The vg gene encodes a nuclear protein expressed during embryogenesis in the presumptive regions of wing and halter imaginal discs in the central nervous system and in a subset of thoracic muscles.7 All vg mutants are characterized by a reduced wing phenotype, indicating a requirement for VG in wing development. During larval development vg expression in the wing disc is dependent on the sequential activation of two intronic enhancers. The boundary enhancer (vgBE) mediates transcription of vg under the control of the Notch pathway via the binding of the suppressor of hairless (Su(H)) transcription factor.4,8 The quadrant enhancer (vgQE) directs vg expression under control of the dpp pathway via the binding of the mothers against dpp (MAD) protein.4,5 In this way, vg integrates inputs from the two axes that pattern wing development.6,9
It has been shown that vg interacts genetically and molecularly with the scalloped (sd) gene product.10,11,12 vg and sd are coexpressed in the wing imaginal disc and dimerization of VG and SD seems to be a prerequisite for translocation of the dimer to the nucleus, where it acts as a functional transcription factor.12,13,14 In fact, in the heterodimer, SD contains the transcription enhancer activator (TEA) DNA binding domain, while VG provides the transcriptional activation domain.10,12,14
It has been shown that ectopic expression of vg in other imaginal discs induces wing outgrowths, this being dependent on the presence of SD.4,11 Therefore, vg seems to be required for both wing identity and cell proliferation, suggesting that the VG–SD dimer could be one of the links between signalling genes and genes involved in cell proliferation.6
We have previously shown a strong interaction between specific antiproliferative drugs, such as methotrexate, aminopterin, fluorodeoxyuridine, and the phenotype of vg mutants.15 These inhibitors of the dihydrofolate reductase (DHFR) and thymidylate synthetase enzymes block DNA replication by depleting the pool of thymidylate (dTMP). This effect can be rescued by the addition of thymidine.16 In addition, several enzymatic activities of the dTMP biosynthesis pathway, such as DHFR and thymidine kinase (TK), are perturbed in vg mutants.15 Therefore, we conclude that vg could be a regulator of dTMP synthesis in the tissue where it is normally expressed.15,17
In mammals, genes involved in dTMP biosynthesis are directly regulated by the E2F transcription factor.18 E2F forms a heterodimer with a subunit encoded by the DP gene family and its expression is required for S-phase transition. In quiescent or differentiated cells, the E2F-DP dimer is bound to the hypophosphorylated form of the retinoblastoma family protein pRB. When cells are stimulated to enter the S phase, the pRb protein is phosphorylated by cyclin-CDK complexes and free E2F-DP factor accumulates, activating genes involved in DNA replication as well as cell cycle progression.19,20 In Drosophila, it has been shown that dE2F1 (called here dE2F) regulates cell cycle progression from G1 to S and its overexpression can drive cells from G2 to M by activating the expression of string (stg), the homolog of yeast cdc25.21,22,23,24,25 Thus, in Drosophila as in mammals, dE2F-1 is a key regulator of cell cycle progression.
Here, we present genetic and molecular data that demonstrate a critical role for vg in cell proliferation and cell cycle progression in the wing imaginal disc. Moreover, vg and sd are able to induce proliferation of HeLa cultured cells. In addition, this indicates that VG–SD targets responsible for inducing proliferation are conserved between humans and Drosophila. Our results indicate that the effects of vg on proliferation are mediated via activation of the expression of dE2F transcription factor and its target genes, RNR2 and stg. However, ectopic vg, but not dE2F, is able to activate DHFR expression in the wing disc, raising the possibility that vg could control genes involved in DNA replication independently of dE2F.
Interestingly, we found that VG and DHFR are, in addition, specifically required for cell survival at the D/V boundary of the wing disc. Using aminopterin, a specific inhibitor of DHFR, we found that a decrease in VG and its target DHFR leads to misregulation of apoptosis regulators such as reaper (rpr) and DIAP1, and subsequent caspase-mediated cell death activation. These results demonstrate that vg is a critical component for both proliferation and cell survival in the wing disc.
Results
vg and sd induce proliferation
vg antagonizes the effect of dacapo
Ectopic expression of vg in different imaginal discs induces wing outgrowth and a role for vg in normal proliferation in the wing disc has been postulated based on the phenotype of vg mutants and vg mutant clonal analysis.4,26 However, other investigators have argued that vg does not induce proliferation but only transdetermination.27,28
To clearly establish the role of vg and sd in proliferation, we overexpressed the Drosophila Dacapo (DAP) protein which shares strong homology with human p21.29 Human p21 has been shown to inhibit cyclin-CDK complexes, Rb phosphorylation and therefore E2F1 factor activation of its target genes. In most cases examined, p21 is essential for G1 arrest before differentiation of cells. In the wing disc, vg is highly expressed along the D/V boundary that will form the future wing margin. We overexpressed dap in this region using the vg-GAL4 driver.30 This led to nicks at the wing tips due to G1 arrest without apoptosis (Figure 1a, b).31 To visualize the effect on proliferation in the wing disc, we analyzed the distribution of M phase cells using the antiphospho-histone H3 mitosis marker. The overexpression of dap led to a significant decrease in mitotic cells along the D/V boundary (Figure 1d, e).
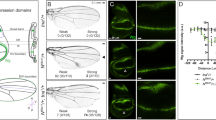
vg antagonises the effect of dap. The vg-GAL4 driver reflects the expression of the vgBE-enhancer. (a) vg-GAL4; vgnull/+ flies display a wild-type wing phenotype. (b) vg-GAL4; UAS-dap; vgnull/+ wing. A scalloped wing phenotype is observed. The same phenotype is observed using the two transgenes: UAS-dap; UAS-LacZ. (c) vg-GAL4; UAS-dap; UAS-vg; vgnull/+ wing. A wild-type phenotype is observed showing that vg antagonises the effect of dap. We assume that this result is not due to titration of the GAL4 protein. M phase staining is correlated with wing phenotypes. Third instar wing discs were stained with antiphospho-histone H3 and anti-β-galactosidase antibodies. All discs are orientated in the same way with the posterior compartment to the right. The genotypes in (d), (e), and (f) correspond to genotypes in (A), (B) and (C) respectively, with an additional vgBE-lacZ insertion that reflects the vg-GAL4 driver at the D/V boundary. A diminution of M cells (arrowheads) is observed at the D/V boundary in (e), while an increase (arrowheads) in staining is detected in (f) explaining the rescue of the wing phenotype observed in (c). The vg-GAL4 driver expression at the D/V boundary is monitored with the vgBE-lacZ staining in (d′), (e′) and (f′), respectively; and merge pictures are presented in (d″), (e″) and (f″)
Next, we expressed both dap and vg using the vg-GAL4 driver and observed a complete rescue of the wing phenotype induced by overexpression of dap (Figure 1c). Consistent with the rescue of the wing phenotype, a significant increase in M phase cells at the level of the D/V boundary of wing imaginal discs was observed when both transgenes were overexpressed compared to when only dap was overexpressed (Figure 1f). Since the role of dap as a general inhibitor of proliferation has been well established, these results strongly argue in favor of a role for vg as a positive regulator of cell proliferation in the zone of the future wing margin.
vg induces G1/S and G2/M phase transitions
Next, we tested whether vg was able to induce proliferation and thus, cell cycle progression (i.e., S and M phases) in other parts of the wing disc using a different driver. In bromodeoxyuridine (BrdU) pulse experiments, we observed significantly fewer cells in the S phase in most of the wing disc in the vgnull mutant than in the wild-type strain (Figure 2a, b). This argument, together with the reduced vgnull disc size, is in favor of a role for vg in cell proliferation. Therefore, the homozygous vgnull genetic context was used in the following experiments, as it emphasizes the effects of ectopic vg on cell cycle transition. As it has already been shown, the ectopic expression of dE2F in the wing disc, using the patched(ptc)-GAL4 driver, is sufficient to drive cells into the S phase (Figure 2d).21 We observed that ectopic expression of vg according to the ptc expression domain in a vgnull background (Figure 2c) also increases the number of cells in the S phase (Figure 2e). Moreover, ectopic vg also led to overgrowth of the disc along the A/P axis (Figure 2c, e). Compared to the wild-type strain, a significant decrease in cells in the M phase was observed in vgnull discs using antiphospho-histone H3 antibodies (Figure 2f, g). Moreover, ectopic expression of vg, driven by ptc-GAL4 in a vgnull background, induced transition to the M phase (Figure 2h). These data indicate that vg is able to regulate proliferation, increasing the number of cells in both the S and M phases. However, we cannot exclude the possibility that the effect of vg on the number of the M phase cells is merely the consequence of induction of the S phase.
vg drives the cell cycle. All discs are orientated in the same way with the posterior compartment to the right. Anti-BrdU antibody marks cells in the S phase in third instar wing disc. (a) Canton-S wild-type disc: all cells are uniformly stained except the zone of nonproliferating cells (ZNC) at the D/V boundary. (b) vgnull wing disc: a decrease in S phase cells in the vgnull mutant is observed. Note the reduced size of the disc and of the wing pouch. (c) Cross between vgnull; UAS-LacZ; UAS-vg and vgnull; ptc-GAL4 flies. β-Galactosidase activity marks the ptc expression domain. Note the overgrowth of the disc along the A/P axis. (d) vgnull disc in which dE2F is overexpressed in the progeny of the cross between vgnull; UAS-E2F-DP; UAS-P35 and vgnull; ptc-GAL4 flies. dE2F1 drives cells to enter the S phase in absence of vg (arrow). The baculovirus P35 protein inhibits dE2F induced cell death. (e) ptc-GAL4-driven ectopic expression of vg in a vgnull background (cross between vgnull; UAS-vg and vgnull; ptc-GAL4 flies). Like dE2F, vg can drive the cell cycle from the G1 to the S phase (arrow). (f) Wild-type wing disc stained with antiphospho-histone H3 antibodies. Uniform staining of cells in the M phase is observed. (g) vgnull wing disc. A strong decrease in staining of the M cells is detected. (h) vgnull wing disc from the F1 of the cross between UAS-vg; vgnull and ptc-GAL4; vgnull flies. Ectopic expression of vg induces mitosis in the ptc expression domain (arrow). (i) Flow cytometry experiments. At 120 h AED (after egg deposition) wing discs were dissociated and analyzed by FACS. en-GAL4; UAS-GFP; UAS-LacZ control disc were compared to en-GAL4; UAS-GFP; UAS-vg discs. Bold and thin traces represent GFP+ (experimental) and GFP− (internal control) cells, respectively. DNA contents data represent the proportions of cells in G1 (2C) and G2 (4C). G1, S and G2 proportions only refer to posterior transgene-expressing cells in the two experiments. Results show that ectopic vg decreases the proportion of cells in G1 and increase the proportion of cells in G2. The experiment was repeated three times. Graphic data come from an experimental set performed in parallel
Ectopic expression of vg changes the repartition of cells in the cycle
To quantify the effect of ectopic expression of vg on the cell cycle, we performed flow cytometry analysis on en-GAL4, UAS-GFP, UAS-vg discs that expresses the green fluorescent protein (GFP) and vg in posterior compartment cells of wing imaginal discs. GFP expression thus allows us to segregate anterior wild-type cells and posterior transgene-expressing cells.24 We compared G1, S and G2/M phases repartition in dissociated wing disc cells, overexpressing vg according to en (GFP+ posterior compartment cells) or not (GFP− anterior internal control compartment). Control en-GAL4, UAS-GFP, UAS-LacZ discs displayed no significant difference between the two compartments (Figure 2i, compare bold and thin lines).24 Conversely, ectopic vg expression truncated the G1 phase, as we observed a 15.15% mean decrease in posterior cells in G1 compared to anterior ones (Figure 2i). These results indicate that ectopic vg causes a shift from the G1 phase to the G2/M phase of the cell cycle and so is an activator of cell proliferation. Furthermore, the analysis of the Forward Scatter data, which is a measure of cell size, reveals no difference in average cell size between the two compartments when vg is overexpressed in the posterior compartment (data not shown). This is surprising as overproliferation is generally accompanied by a reduction in cell size. However, this could be explained if VG not only activates cell proliferation, but also promotes cell growth. These data and those of the former paragraph confirm that vg is able to drive the cell cycle and induce proliferation in the wing disc.
VG and SD induce proliferation of HeLa cells
Our data show that ectopically expressed vg induces proliferation in vivo. In order to evaluate if the capacity of the VG–SD dimer, per se, to induce proliferation is dependent on other components expressed in the wing disc, we tested whether the dimer can induce proliferation in cultured cells. In addition, we chose to transfect vg and sd constructs into heterologous HeLa cells that are devoid of Drosophila proteins. The rate of proliferation was estimated by measuring mitochondrial dehydrogenase enzyme activity.32,33
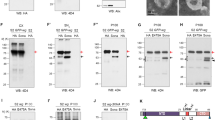
We observed that the cotransfection of vg and sd induces a 2.5-fold increase in the proliferation rate of HeLa cells (Figure 3). In accordance with the model in which VG acts as a transcription activator and SD carries only the DNA binding domain, we found that transfection of vg alone did not significantly induce cell proliferation. Interestingly, the transfection of sd alone seemed to decrease the rate of proliferation, suggesting that SD acts in a dominant-negative fashion on endogenous human proteins implicated in this process (Figure 3). These results indicate that the VG–SD dimer is able to induce proliferation of cells in culture per se. Moreover, it suggests that VG–SD targets implicated in cell proliferation are conserved between Drosophila and human.
VG and SD induce cell proliferation. Cell transfection. vg, sd and GFP cDNAs containing plasmids were transfected into HeLa cells. A quantity of 500 ng per plasmid was transfected. GFP-positive cells were sorted by flow cytometry, collected and six different samples were cultured for each transfection. Proliferation was quantified by spectrophotometric measure of a mitochondrial dehydrogenase activity. Mean values were obtained. White: control transfection of the mock plasmid devoid of cDNA. Dark grey: transfection of vg alone. Some induction of cell proliferation is visible. Black: transfection of vg and sd. A 2.5-fold induction of proliferation is observed. Light grey: transfection of sd only. Inhibition of proliferation is observed suggesting a dominant-negative effect. Each set of experiments was independently performed three times at least. Standard deviation are shown
Ectopic vg expression induces dE2F and dE2F target genes
vg induces dE2F expression
A prerequisite for cell proliferation is the activation of genes encoding products required for DNA replication, as well as genes involved in regulation of the cell cycle. In mammals, the E2F transcription factor regulates the expression of genes involved in DNA replication, like DHFR, ribonucleotide reductase (RNR2) and TK. Two dE2F genes have been identified in Drosophila, dE2F1 and dE2F2.23,25,34 dE2F1 (named here dE2F) has been shown to control RNR2 and proliferating cell nuclear antigen (PCNA) gene expression, while a role for dE2F in DHFR and TK activation has not been investigated yet.22,35
A possible link between vg and dE2F was investigated first by testing for genetic interactions in double heterozygous mutant combinations of a dE2F null allele (dE2F91),22 and either vg or sd mutations. No interaction was observed in vgnull and dE2F91 double heterozygotes (data not shown). However, vg is strongly haplo-sufficient and double heterozygous sd58/+; vgnull/+ females displayed a wild-type wing phenotype although sd is a well-established partner of vg. Only sdETX4/Y males displayed an enhanced wing phenotype in a vgnull/+ heterozygous background (Figure 4a, b).11 Similarly, sdETX4/Y, dE2F91/+ males displayed a more severe mutant wing phenotype compared to sdETX4/Y males (Figure 4a, c), suggesting a genetic interaction between dE2F and sd.
sd and dE2F interact genetically. (a) Wing of a sdETX4/Y male. (b) wing of a sdETX4/Y; vgnull/+ mutant. The increase in the mutant wing phenotype in a vgnull heterozygous background reflects the known protein–protein interaction between the two products. (c) Wing of a sdETX4/Y; dE2F91/+ mutant. As in (b), an increase in the mutant phenotype is also observed. Ectopic expression of vg induces dE2F expression. All discs are orientated in the same way with the posterior compartment to the right. (d) In situ hybridization with a dE2F antisense RNA probe on a wild-type third instar wing disc. (e) Third instar wing disc of progeny of the cross between UAS-vg and en-GAL4 strains. vg is overexpressed in the posterior compartment. An induction of dE2F is observed. (f) Third instar wing disc of progeny of the cross between UAS-vg and ptc-GAL4; E2F-LacZ strains. A faint expression of dE2F is normally observed at the level of the D/V boundary. The arrow shows ectopic expression of β-galactosidase according to ptc expresssion pattern confirming the in situ experiment observed in (e). (g) vgnull wing disc in which vg is overexpressed in the progeny of the cross between vgnull; UAS-vg and vgnull; ptc-GAL4 flies. Staining with anti-dE2F antibodies reveals that vg induces the expression of the dE2F protein
We therefore examined the hypothesis that VG–SD induces cell proliferation through regulation of the dE2F transcription factor. Since it is not possible to obtain vgnull clones and since the wing pouch is severely reduced in the vgnull mutant, we used the overexpression UAS-GAL4 system to address this issue.4,30,36 We first tested if vg is able to induce dE2F ectopically, using in situ hybridization with RNA probes. In the wild-type disc, dE2F is expressed at relatively low levels in the notum and the wing pouch (Figure 4d). However, it has already been shown that dE2F, though expressed at the D/V boundary, is lately inactive.22,37
We overexpressed vg in the entire posterior compartment of the wing disc using the en-GAL4 driver. Under these conditions, an increase in dE2F RNA expression was observed in the posterior part of the wing disc (Figure 4e). These results were confirmed at the protein level by immunostaining using anti-dE2F antibodies when we overexpressed vg in the ptc domain (Figure 4g). The E2Frm729 mutant is a P[LacZ] element inserted in the dE2F gene.22 In ptc-GAL4, UAS-vg, E2F-LacZ flies, β-galactosidase activity was observed in the ptc expression domain, suggesting that the VG–SD dimer acts on the dE2F promoter (Figure 4f). However, we cannot exclude the possibility that VG–SD induces expression of other targets that would in turn activate the dE2F promoter.
vg induces dRNR2 and stg
In mammals, E2F1 activates a large variety of genes involved in DNA replication, cell cycle progression and oncogenesis. In mammals, the gene encoding RNR2 is regulated by E2F1, and in Drosophila by dE2F1.22 Moreover, it has been shown that, in Drosophila, overexpression of dE2F directs not only cell cycle progression from G1 to S, but also from G2 to M by activating the expression of the stg gene, a homolog of yeast cdc25.24 We wanted to determine if DmRNR2 and stg are also vg targets. In situ hybridization with a DmRNR2 probe revealed expression of this gene in the wing disc but not at the level of the wing margin (Figure 5a). Johnston and Edgar37 hypothesized that the absence of DmRNR2 expression at the wing margin is due to the fact that dE2F1 is probably inactive since these cells are arrested in G1.
Ectopic vg induce DmRNR2, stg and DHFR. In situ hybridization with antisense RNA probes on third instar wing discs. All discs are orientated in the same way with the posterior compartment to the right. Wild-type discs (a, c, e) are compared to third instar wing discs of progeny from the cross between UAS-vg and en-GAL4 strains (b, d, f) in which vg is overexpressed in the posterior compartment. The probes used were: DmRNR2 (a, b), stg (c, d) and DHFR (e, f). All three genes are overexpressed in the posterior compartment in en expression domain, in response to ectopic vg
We ectopically expressed vg in the posterior compartment of the wing disc using the en-GAL4 driver. Strong DmRNR2 expression was induced in this posterior compartment compared to the control (Figure 5a, b), indicating that vg can induce this identified target of dE2F. In the same way, stg expression was detected in wild-type wing discs as previously described,1 and was induced by ectopic vg (Figure 5c, d). Taken together our results show that ectopic vg is sufficient to induce expression of a functional dE2F transcription factor and two of its target genes dmRNR2 and stg.
vg, but not dE2F, induces DHFR
Another well-identified target of E2F1 in mammals is the DHFR gene. DHFR is involved in a critical step of dTMP synthesis and, thus, is necessary for DNA replication. In situ hybridization with a Drosophila DHFR probe showed almost uniform staining in the wild-type wing disc (Figure 5e). It had not been established whether dE2F is able to induce DHFR expression in Drosophila. Surprisingly, we found that ectopic expression of dE2F using the en-GAL4 driver had no effect on DHFR expression in the wing disc (data not shown). Since ectopic expression of dE2F induces apoptosis, we coexpressed dE2F and the baculovirus P35 protein which inhibits cell death.21 No induction of DHFR either was observed even under these conditions (data not shown).
We next tested by in situ hybridization whether overexpression of vg in the en expression domain of the wing disc was able to induce DHFR expression. Indeed vg induced strong DHFR expression in the posterior part of the wing disc, the effect being particularly striking in the wing pouch (Figure 5e, f).
Genetic interactions between vg, but not dE2F, and def(DHFR)
To test, at a genetic level, the interaction between vg and DHFR, we used two different DHFR deficiencies since no DHFR mutants were available (see Materials and Methods). As for vg and dE2F, no interaction was observed between vg and def(DHFR) in double heterozygotes (vgnull/+; def(DHFR)/+) (data not shown). However, the wing phenotype of sdETX4/Y mutants is severely aggravated in a def(DHFR)/+ context (Figure 6a, b), indicating a genetic interaction between DHFR and sd. We cannot exclude at this point the possibility that this interaction is due to other genes included in the deficiency even if no likely candidate genes could be identified in the database.
Genetic interaction between sd and DHFR. (a) Wing of a sdETX4/Y male. (b) Wing of a sdETX4/Y; def(DHFR)/+ male. A strong increase of the mutant phenotype is observed in a def(DHFR) heterozygous background, suggesting a genetic interaction between the two genes. Genetic interaction between vg and DHFR in the presence of a low dose of aminopterin (0.5 mg/kg). Each picture shows the most representative phenotype of the cross. (c) vgnull/+ wing. A scalloped phenotype is observed. (d) vgnull/+; def(DHFR)/+ wing. A strong increase in the mutant phenotype is observed compared to (c). (e) dE2F91/+; vgnull/+ wing. Small notches at the wing tips are observed. (f) Wing of the wild-type strain which also corresponds to what is observed for def(DHFR)/+ or dE2F91/+; def(DHFR)/+ flies
In order to try to reveal possible genetic interactions between vg, dE2F and DHFR, we used very low doses of aminopterin (0.5 mg/kg) to reduce the strong haplo-sufficiency associated with vg mutations (see the section ‘vg induces dE2F expression’). Indeed aminopterin is a specific inhibitor of DHFR activity and it has been shown to lead to a decrease in vg expression in wild-type strains.17 When flies were reared on a low dose of aminopterin (0.5 mg/kg), the slight inhibition of DHFR activity led to nicks in wings in 2% of def(DHFR)/+ heterozygous flies (data not shown). Potential genetic interactions between dE2F and def(DHFR) or vg and def(DHFR) should lead to increased wing phenotypes in double heterozygous flies reared on the drug, compared to def(DHFR)/+ controls. Wild type or dE2F91/+ heterozygous flies exhibit no wing abnormalities when raised at this low concentration of aminopterin (Figure 6f). Nicks at wing tips were observed in vgnull/+ heterozygous flies reared on aminopterin (Figure 6c), while a much stronger notched wing phenotype was obtained in vg/+; def(DHFR)/+ double heterozygotes (Figure 6d). No differences in wing phenotypes were detected in vgnull/+; E2F91/+ (Figure 6e) or in the E2F91/+; def(DHFR)/+ double heterozygotes flies reared on the inhibitor (Figure 6f and data not shown) compared to the controls.
These results demonstrate a strong genetic interaction between vg and def(DHFR) leading to a wing margin phenotype. In contrast, no interaction was detected between dE2F and vg or dE2F and def(DHFR).
These results suggest that in the Drosophila wing disc, unlike what has been described in mammals, dE2F does not control DHFR expression, this role possibly being allotted to the VG–SD dimer. This raises the possibility that in addition to DHFR, other identified targets of dE2F may also be regulated in Drosophila by transcription factors involved in proliferation, others than dE2F.
Diminution of DHFR activity leads to D/V boundary defects
In order to distinguish the respective roles of DHFR and dE2F at the D/V boundary of the wing disc, we used the cut-LacZ strain, which precisely marks a stripe of D/V boundary cells and allowed to monitor their fate. To reduce DHFR activity we also used aminopterin at a concentration of 2 mg/kg to correlate the wing phenotype and D/V boundary cell fate in different genotypes. No difference in the cut-LacZ expression pattern was detected between wild-type flies and vgnull/+ heterozygous flies reared on normal medium (Figure 7a). Nevertheless, in flies grown on medium containing aminopterin, although the wild-type strain exhibited normal cut-LacZ expression (Figure 7b), vgnull/+ heterozygotes displayed significant gaps in the staining (Figure 7c). Moreover, these discs also displayed a high level of cell death specifically at the level of D/V boundary that could be visualized by staining with acridine orange (Figure 7k). No other part of this disc was exposed to such a specific increase in cell death. This was not observed in the wild-type strain where quite homogenous cell death in the wing disc was detected (Figure 7j). As aminopterin is a specific inhibitor of DHFR activity, these results suggest that a decrease in DHFR in a vgnull/+ heterozygous mutant background induces specific cell death at the D/V boundary and thus loss of cut-LacZ-expressing cells.
vg together with DHFR are required for cell survival at the D/V boundary of the wing imaginal disc. Effect of aminopterin, the inhibitor of DHFR. All discs are orientated in the same way with the posterior compartment to the right. X-gal staining was performed rigorously, with the same experimental procedure for all genotypes. Staining times were precisely the same and experiments were repeated four times. We could not find any difference in staining between the wild-type Canton S and w1118 strain, which is the genetic background for the different mutants. The cut-LacZ transgene marks a stripe of cells precisely at the D/V boundary and allows to monitor their fate (a–e). (a) Wild-type wing disc heterozygous for the cut-LacZ transgene. Similar LacZ expression is observed in a vgnull/+ heterozygous disc (data not shown). (b) Wild-type wing disc of flies reared on medium containing a concentration of 2 mg/kg of aminopterin. cut-LacZ pattern is normal suggesting absence of D/V boundary specific apoptosis in LacZ-expressing cells. (c) In vgnull/+ heterozygotes, LacZ staining at the D/V boundary is interrupted, probably due to diminution of DHFR activity induced by aminopterin (arrows). (d) No visible effect is observed in E2F91/+ wing discs. (e) Conversely, in a def(DHFR)/+ background, gaps at the level of the D/V boundary are observed (arrows). The expression of the apoptosis inducer gene reaper (rpr) was monitored using the rpr-LacZ transgene. Flies were reared on medium containing aminopterin (0.5 mg/kg) (f–i). Aminopterin has no effect on rpr expression in the canton S wild-type strain (f and data not shown), which is normally detected in the wing pouch, particularly at the D/V boundary. (g) Conversely, in vgnull/+ heterozygous wing discs, the rpr-LacZ reporter construct shows an increased activity especially at the D/V boundary. (h) E2F91/+ wing discs do not display D/V boundary specific rpr-lacZ activation (a slight decrease can even sometimes be observed). (i) def(DHFR)/+ discs resemble the vgnull/+ context. This specific rpr-lacZ induction correlates with the disappearance of D/V boundary cut-LacZ-expressing cell reported (c, e). Cell death was monitored with acridine orange staining. (j) Aminopterin (0.5 mg/kg) induces apoptosis quite homogeneously in wild-type wing discs. (k) Conversely, a specific increase in apoptosis at the D/V boundary is visualized in vgnull/+ heterozygous discs
When reared on aminopterin, def(DHFR)/+ discs clearly displayed abnormal cut-LacZ staining (Figure 7e) like that in vgnull/+ heterozygotes (Figure 7c). This is in agreement with the hypothesis that the abnormal cut-LacZ phenotype is due to a decrease in DHFR activity.
If the DHFR gene was to be regulated by dE2F at the D/V boundary, then reduced DHFR expression in dE2F91/+ mutant wing discs and consequent abnormal cut-LacZ expression in larvae reared on the inhibitor should be expected. This was not the case since cut-LacZ expression was normal in this genetic background (Figure 7d). This is in accordance with the hypothesis that vg, but not dE2F, is able to regulate DHFR expression at the D/V boundary in wing discs of the third instar larvae. Moreover, these results suggest that VG is also required for cell survival in the wing disc.
vg and DHFR are specifically required for cell survival at the D/V boundary
We next tested whether the effects observed on wing margin cell fates and cell death induction, when flies of different genotypes were reared on aminopterin (0.5 mg/kg), could be correlated with misregulation of apoptosis regulators. Drosophila inhibitor of apotosis protein 1 (DIAP1) is a negative regulator of apoptosis that binds to and inhibits caspase activity.38 DIAP1 has recently been reported to be detected throughout the wing disc with an increased expression pattern along the pouch D/V boundary.39 The authors also demonstrated that the proapoptotic Reaper (Rpr) protein is able to bind to and trigger ubiquitin-mediated DIAP1 degradation leading to subsequent caspase activation and apoptosis.39 But more concerning is the fact that rpr is also expressed in the wing pouch, particularly at the D/V boundary.40,41 This boundary, therefore, seems to be a quite sensitive area under tight regulation of cell death and cell survival signals. To monitor rpr expression in the different vg, dE2F and DHFR heterozygous contexts, we used a rpr-LacZ reporter transgenic line.41,42 No difference in LacZ expression was observed between the different genotypes when flies were reared on normal medium (data not shown). On aminopterin-containing medium (0.5 mg/kg), specific induction of rpr was not observed at the level of the wing margin in the wild-type strain (Figure 7f and data not shown) nor in dE2F91 heterozygotes (Figure 7h) where a slight decrease in rpr-LacZ expression at the D/V boundary can even sometimes be observed. Conversely, rpr was specifically activated at the D/V boundary in vgnull/+ (Figure 7g) and def(DHFR)/+ (Figure 7i) heterozygotes, indicating a complete correlation with the loss of cut-LacZ-expressing cells and cell death induction.
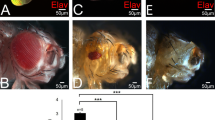
These results led us to investigate the effect of rpr induction on DIAP1, upon reduction of VG and DHFR levels. Strikingly, DIAP1 protein was specifically and severely downregulated at the D/V boundary of vgnull/+ mutants compared to wild-type discs (Figure 8a, b). Nevertheless, no effect could be observed on DIAP1 transcription, using a P[LacZ] insertion at the thread (th) locus encoding DIAP1 (Figure 8d and data not shown). These results are consistent with the rpr-induced post-translational degradation of DIAP-1 reported so far. Conversely, vg overexpression along the D/V boundary (vg-GAL4 driver) completely rescued the loss of DIAP1 in vgnull/+ disc (Figure 8c), as expected if DIAP1 is regulated by VG. We concluded that vg requirement for cell survival at the D/V boundary is, at least, mediated through DIAP1.
Apotosis inhibitor DIAP1 is required for regulation of cell survival at the D/V boundary of the wing disc by vg. All discs are orientated in the same way with the posterior compartment to the right and ventral part on the top. (a) anti-DIAP1 staining. DIAP1 protein is observed throughout the whole wing disc of wild-type strain with a striking increase in expression at the D/V boundary (arrowheads), as already described.39 (b) This D/V specific DIAP1 protein is lost in vgnull/+ heterozygous mutants (arrowheads). (c) vg overexpression along the D/V boundary (vgGAL4 driver) of vgnull/+ disc completely rescues DIAP1 protein level (arrowheads). (d) anti-β-galactosidase staining. P[LacZ] insertion at the th locus encoding DIAP1 reflects diap1 transcription with a marked increase at the D/V boundary of wild-type disc (arrowheads).39 No difference can be monitored in a vgnull/+ context (data not shown). (e) Addition of aminopterin (0.5 mg/kg) to flies medium leads to DIAP1 downregulation similar to (b) (arrowheads). (f) Moreover, vg overexpression along the D/V boundary rescues DIAP1 protein, as well (arrowheads). (g) CM1 antiactivated caspases staining of wild-type disc. Positive cells are quite rare in the wing pouch (arrowheads). (h) Only few caspase-positive cells are observed in vgnull/+ disc (arrowheads and inset). (i) Same disc stained with DAPI. Caspase-positive cells display pyknotic nuclei (arrowheads and inset). (j) Upon addition of aminopterin (0.5 mg/kg), caspase-positive cells are observed in a quite homogeneous pattern in the wing disc. (k) Strikingly, activation of caspases specifically increases at the D/V boundary of vgnull/+ discs gown on aminopterin (arrowheads and inset). (l) Same disc staining with DAPI. Pyknotic nuclei are particularly observed along the D/V boundary (arrowheads and inset). (m) DIAP1 overexpression along the D/V boundary (vgGAL4 driver) rescues to some extent the wing phenotype of vgnull/+ flies grown on aminopterin (0.5 mg/kg)
Interestingly, specific inhibition of DHFR (aminopterin 0.5 mg/kg) in wild-type flies also induced DIAP1 downregulation (Figure 8e) that was rescued upon vg overexpression (Figure 8f). These results confirm that a decrease in the VG product shifts the D/V boundary cells to a cell death sensitive state that seems to be correlated with a reduced DHFR activity.
Finally, we monitored the effects of aminopterin on cell death effectors, using the CM1 antiactivated caspases antibody. Only few caspase-positive cells were detected in wild-type or vgnull/+ discs grown on normal medium (Figure 8g, h). These cells displayed pyknotic nuclei, indicative of apoptosis (Figure 8h, i).43 However, we observed that vgnull/+ discs on aminopterin displayed a high level of activated caspases, which was especially restricted to the D/V boundary (Figure 8k) compared to wild-type ones (Figure 8i). Many vgnull/+ cells along the D/V boundary displayed pyknotic nuclei (Figure 8k, l). This result accounts for acridine stained cell death that was previously observed (Figure 7k), and is consistent with caspase inhibitor DIAP1 downregulation we report.
Moreover, DIAP1 overexpression at the D/V boundary (vgGAL4 driver) greatly rescued the strong wing margin phenotype observed in vgnull/+ flies grown on aminopterin (0.5 mg/kg) (Figure 8m). These results demonstrate that DIAP1 degradation stands for caspase-mediated apoptosis induced in vgnull/+ discs grown on aminopterin (Figure 7).
In these experiments too, the data show that DHFR activity is required for cell survival at the D/V boundary and would be under the control of vg. A decrease in VG or DHFR activity would lead to specific rpr expression, DIAP1 degradation, caspase-mediated cell death and, consequently, to wing margin defects.
Discussion
The normal development of multicellular organisms requires proliferation of precursor cells followed by cell cycle arrest and commitment to specialized fates. In mammals, E2F, P21 and the pRb pocket proteins have been shown to be involved in differentiation.20 However, little is known about how developmental pathways regulate the cell cycle. Regulation of p21 by HOX genes has been reported in a human myelomonocytic cell line; this constitutes one of the few examples described so far.44 Recently, the transcription factor Cubitus interruptus (Ci) was shown to mediate the effects of Hedgehog (Hh) signalling on cell growth and proliferation in the wing disc by direct binding to the Cyclin E promoter.45 In addition, the Caudal (CAD) homeodomain protein was shown to bind the dE2F promoter in vitro, and CAD binding sites in the dE2F promoter are required for expression of dE2F in living flies.46 These results most clearly demonstrate that developmental genes like Cad, Hh and Hox can directly regulate cell growth and proliferation.
There are several indications that the vg gene can induce cell proliferation when expressed ectopically,4,47 and may mediate the cross-talk between developmental and cell cycle genes in wing development. Interestingly, Notch and dpp signalling pathways have been shown to both activate vg in the wing disc via specific enhancers, and induce proliferation. In a recent report, cell proliferation induced by ectopic expression of an activated form of Thick vein (TKV), the DPP receptor, was shown to require the presence of VG. However, TKV was unable to induce vg expression, indicating only a synergetic effect between the two genes in this process.48
The results presented here clearly demonstrate that vg is required and sufficient for cell proliferation at the D/V boundary of the wing imaginal disc. Not only can the overexpression of the gene counteract the effect of dap, but it also induces cell cycle progression. The latter was confirmed by: (i) overexpression of vg in the wing disc and consequent induction of G1/S and G2/M cell progression in the region where vg was overexpressed and (ii) fluorescence-activated cell sorter (FACS) experiments showing that overexpression of vg in the posterior compartment of the wing disc shifts the cells toward the G2/M phase. VG therefore seems to be an active regulator of G1/S phase transition.
Moreover, in a heterologous system (HeLa cells) the Drosophila VG–SD dimer, per se, induces cell proliferation. This suggests that the dimer is able to act on human proliferation genes and, thus, that target sites of the SD TEA DNA binding domain are likely conserved between Drosophila and humans. In addition, our results in human HeLa cells are in favor of a dominant-negative effect of an excessive amount of SD, which probably saturates the target sites involved in the process and results in inhibition of cell proliferation. On the whole, data are in favor of the hypothesis that the target sites of the SD TEA DNA binding domain are conserved in Drosophila and human and are necessary for cell proliferation.
The prerequisite for cell proliferation is the activation of specific transcription factors, like E2F, that trigger DNA replication and cell cycle progression. Our results show that vg induces cell proliferation in the wing disc by activating dE2F, but also dE2F target genes like dmRNR2 and stg.
Although DHFR is a well-established target of dE2F in mammals, we found that in contrast to dE2F, vg induces DHFR expression in the Drosophila wing imaginal disc, suggesting the possibility that vg exerts some of its effects directly on proliferation genes in the wing pouch and not merely through dE2F. In addition, on medium containing low concentration of aminopterin that inhibits DHFR activity, a genetic interaction was observed between vg and def(DHFR), and not between the latter and dE2F, with respect to wing phenotype.
Moreover, the genetic interaction between vg and def(DHFR) leading to wing margin phenotypes was corroborated with the loss of D/V boundary cut-LacZ-expressing cells in vgnull/+ and def(DHFR)/+ heterozygotes grown on aminopterin. No such effect could be visualized in dE2F91/+ mutant, confirming that the regulation of DHFR by vg was not mediated by dE2F.
We further showed that vgnull/+ mutants on aminopterin displayed a clear increase in cell death that was restricted to the D/V boundary. These results led us to monitor a potential additional role for vg in cell survival. Interestingly, the strong expressions of both apoptosis regulators rpr and DIAP1, specifically at the D/V boundary of the wing disc, are in favor of a precise regulation of cell survival in this area.40,41 In addition, the fact that vg is highly expressed in the same area and is absolutely required for wing pouch development, led us to monitor cell death regulators expression in both wild-type and vgnull/+ discs.7 Using aminopterin, we showed that VG and DHFR are required for the integrity of wing margin cells, in addition to cell proliferation. A decrease in both products, induces specific rpr expression, DIAP1 downregulation and caspase-mediated cell death at the D/V boundary of the wing disc. However, even if DIAP1 was specifically lost in vgnull/+ cells at the D/V boundary, we could not detect any clear induction of apoptosis without addition of aminopterin. This indicates that VG reduction only promotes a cell death sensitive state. Apoptosis was further triggered upon concomitant DHFR inhibition. However, it has been shown that vg displays incomplete penetrance depending on genetic backgrounds.49 This suggests that cell death eventually occurs in heterozygotes on normal medium.
Interestingly, our results clearly show that vg and its target DHFR are required for both cell survival and proliferation. Therefore, the complete loss of wing tissue in vgnull homozygous mutant7,11 and the impossibility to recover vgnull clones4,36 are probably due to both proliferation impairment and apoptosis triggering. The requirement for DHFR during both processes raises the possibility of mutual cross-talk between cell survival and proliferation that would rely on VG function and DNA synthesis. Moreover, the wing phenotype of vgnull/+ flies grown on aminopterin was greatly rescued upon ectopic expression of DIAP1 along the D/V boundary. This demonstrates that wing defects observed upon DHFR inhibition and VG reduction are significantly due to cell death triggering.
It has been proposed that vg can be viewed as a wing selector gene that would impose the wing developmental pathway.27,50,51 Interactions between vg and genes like Notch, wg and dpp would influence gene expression patterns to promote wing identity.28 However, some authors recently concluded that vg is not a ‘master gene’ but a component of the genetic combination that drives wing blade development. Thus, only an indirect role of vg in cell proliferation was hypothesized by these authors.52 In this study, we demonstrate that vg not only promotes wing differentiation, but is also required for cell survival and proliferation.
In addition, several counter arguments can be used to uncouple a role for vg in cell proliferation from its role in cell identity. Indeed, TONDU (TDU) a human vg homolog, dimerizes with SD when expressed in flies.14 Ectopic expression of TDU in the eye disc induces cell proliferation without promoting wing cell differentiation.14 Another VG partner in the wing disc has been identified, Strawberry notch (SNO), which is also a Notch target.53,54 The possibility exists that wing specification requires more than just the VG–SD transcription factor, while the VG–SD dimer alone would be sufficient to induce proliferation, as was suggested by our experiments of heterologous expression of vg and sd in HeLa cells. Indeed, it has been shown that SD DNA target selectivity can be modified when SD is bound to VG.55 In vitro studies on the binding of the TEA domain of SD on well-established targets show that the presence of a cofactor, in this occurrence VG, dramatically shifts the affinity of SD for these targets. Therefore, we can hypothesize that induction of SD target genes depends on different components bound to the SD protein.
Taken together our results point to the existence of a tissue-specific activator of dE2F and DHFR, namely vg, which is required for cell proliferation and cell survival. On the whole, our results demonstrate that vg drives cell cycle progression in the wing disc, acting on the expression of key genes that control the pathway, and that the VG–SD dimer retains its potentialities as an inductor of cell proliferation in a heterologous system. Moreover, we show that vg is also required for normal expression of apoptosis regulators and cell survival in the presumptive wing margin area.
Materials and Methods
Drosophila strains and medium
The vgnull and UAS-vg strains were generated in our laboratory.11 The vg-GAL4 strain that drives GAL4 expression according to part of the vgBE-enhancer was provided by M Hoffmann and S Morimura. The vgBE-lacZ strain that drives β-galactosidase expression according to the vgBE enhancer was described in Williams et al.8 The sdETX4 strain is a sd-LacZ enhancer trap described in Campbell et al.13 The E2Frm729 line, which contains an enhancer trap inserted in the dE2F1 gene, is called E2F-lacZ in this study.22 The E2F91 mutation is described as a null allele for the gene.22 The en-GAL4; UAS-GFP (GFP) strain was used for FACS experiments. The DHFR gene is located at 89D2. The first deficiency used is Df (3R) p10 (Bloomington no. 3483), (breaks at 89C1–89E2). UAS-dap is a gift from C Lehner and UAS-E2F-DP; UAS-P35 a gift from B Edgar. The latter strain allows expression of dE2F without induction of cell death. The rpr-LacZ strain that carries the lacZ sequence under the control of the entire promoter of the rpr gene was provided by JM Abrams.41,42 The thj5c8 strain that carry a P[lacZ] reporter in the 5′ untranslated region of the diap1 transcription unit and the UAS-diap1 strain were provided by K White and H Steller.39 All other stocks come from the Bloomington Drosophila Center. Aminopterin (Sigma) was added to the medium at a concentration of 0.5 or 2 mg/kg depending on the experiment.
Histology
A Leica DMR microscope was used. X-Gal staining was performed as described31 and viewed using Nomarski optics. Digoxigenin (DIG)-labeled antisense or sense RNA probes were generated with T3 or T7 RNA polymerase (Promega) and DIG-UTP (Roche) from cloned cDNAs of dE2F1, DHFR, stg and DmRNR2. These probes were used for whole-mount in situ hybridization of fixed larval imaginal discs. The DIG-labeled RNA probes were detected with the aid of an anti-DIG antibody coupled to alkaline phosphatase (Roche) and NBT/BCIP as substrate. BrdU and DAPI labeling of imaginal discs was performed as described.1 Staining with rabbit antiphospho-histone H3 antibody (Upstate Biotechnology), anti-VG (gift from S Carroll), anti-dE2F1 (gift from P Leopold) anti-DIAP1 (gift from HD Ryoo and H Steller), CM1 antiactivated caspases antibodies (BD Pharmingen) and mouse anti-β-galactosidase antibody (Jackson Immuno Labs) were performed according to standard protocols.
FACS experiments
Staged larvae (120 h after egg deposition) derived from 2 to 3 h egg collections and raised at 25°C were dissected in PBS for each genotype. In all, 20 wing discs were washed twice in PBS and incubated with gentle agitation in trypsin (Invitrogen) and Hoechst 33342 (Sigma) for 150 min according to Nuefeld et al.24 About 20 000 GFP+ and GFP− cells were analyzed for each genotype. An Elite Beckman Coulter FACS was used and data were analyzed using the Multicycle Software. P-values were calculated using a two-tailed Student's t-test.
Cell transfection
vg and sd cDNAs were subcloned in the pXJ40 vector (gift from I Davidson) in order to allow expression of these cDNAs under the control of a CMV promoter. HeLa cells were transfected using the FuGENE6™ reagent (Roche), with pXJ40 plasmids containing vg, sd cDNAs and pEGFP (GFP) (Clontech) according to the supplier's instructions.
Cell proliferation
Transfected HeLa cells were incubated for 48 h at 37°C in an atmosphere containing 5% CO2. GFP-positive cells were sorted by flow cytometry (Fluorescent Activated Cell Sorter FACS), collected, and then placed in 96-well plates (2000 cells per well) and cultured for 48 h at 37°C in 100 μl of medium. In all, 50 μl of coloring solution XTT (0.3 mg/ml) (Cell proliferation kit II, Roche), which allows quantification of cell proliferation by measuring mitochondrial dehydrogenase activity, was added to each well.32,33 Light Absorption in each well was measured at 490 and 640 nm 4 and 48 h later with an ELISA reader (MRX II Thermosystem).
Abbreviations
- vg :
-
vestigial
- sd :
-
scalloped
- DHFR:
-
dihydrofolate reductase
- vgBE :
-
boundary enhancer
- Su(H):
-
suppressor of hairless
- MAD:
-
mothers against dpp
- TEA:
-
transcription enhancer activator
References
Milan M, Campuzano S and Garcia-Bellido A (1996) Cell cycling and patterned cell proliferation in the wing primordium of Drosophila. Proc. Natl. Acad. Sci. USA 93: 640–645
Blair SS (1995) Compartments and appendage development in Drosophila. Bioessays 17: 299–309
de Celis JF (1999) The function of vestigial in Drosophila wing development: how are tissue-specific responses to signalling pathways specified? Bioessays 21: 542–545
Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J and Carroll SB (1996) Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature 382: 133–138
Kim J, Magee J and Carroll SB (1997) Intercompartmental signaling and the regulation of vestigial expression at the dorsoventral boundary of the developing Drosophila wing. Cold Spring Harb. Symp. Quant. Biol. 62: 283–291
Cohen SM (1996) Controlling growth of the wing: vestigial integrates signals from the compartment boundaries. Bioessays 18: 855–858
Williams JA, Bell JB and Carroll SB (1991) Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes Dev. 5: 2481–2495
Williams JA, Paddock SW, Vorwerk K and Carroll SB (1994) Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature 368: 299–305
Neumann CJ and Cohen SM (1996) A hierarchy of cross-regulation involving Notch, wingless, vestigial and cut organizes the dorsal/ventral axis of the Drosophila wing. Development 22: 3477–3485
Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD and Bell JB (1998) Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 12: 3815–3820
Paumard-Rigal S, Zider A, Vaudin P and Silber J (1998) Specific interactions between vestigial and scalloped are required to promote wing tissue proliferation in Drosophila melanogaster. Dev. Genes Evol. 208: 440–446
Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A and Carroll S (1998) The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 12: 3900–3909
Campbell S, Inamdar M, Rodrigues V, Raghavan V, Palazzolo M and Chovnick A (1992) The scalloped gene encodes a novel, evolutionarily conserved transcription factor required for sensory organ differentiation in Drosophila. Genes Dev. 6: 367–379
Vaudin P, Delanoue R, Davidson I, Silber J and Zider A (1999) TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development 126: 4807–4816
Silber J, Bazin C and Le Menn A (1989) Vestigial mutants of Drosophila melanogaster live better in the presence of aminopterin: increased level of dihydrofolate reductase in a mutant. Mol. Gen. Genet. 218: 475–480
Silber J, Le Menn A, Chevillard S, Zider A and Paumard S (1993) The vestigial locus of Drosophila melanogaster is involved in resistance to inhibitors of dTMP synthesis. Mol. Gen. Genet. 241: 42–48
Zider A, Flagiello D, Frouin I and Silber J (1996) Vestigial gene expression in Drosophila melanogaster is modulated by the dTMP pool. Mol. Gen. Genet. 251: 91–98
Johnson DG, Schwarz JK, Cress WD and Nevins JR (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature 365: 349–352
Harbour JW and Dean DC (2000) The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14: 2393–2409
Dyson N (1998) The regulation of E2F by pRB-family proteins. Genes Dev. 12: 2245–2262
Asano M, Nevins JR and Wharton RP (1996) Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 10: 1422–1432
Duronio RJ, O'Farrell PH, Xie JE, Brook A and Dyson N (1995) The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9: 1445–1455
Dynlacht BD, Brook A, Dembski M, Yenush L and Dyson N (1994) DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc. Natl. Acad. Sci. USA 91: 6359–6363
Neufeld TP, de la Cruz AF, Johnston LA and Edgar BA (1998) Coordination of growth and cell division in the Drosophila wing. Cell 93: 1183–1193
Ohtani K and Nevins JR (1994) Functional properties of a Drosophila homolog of the E2F1 gene. Mol. Cell. Biol. 14: 1603–1612
Go MJ, Eastman DS and Artavanis-Tsakonas S (1998) Cell proliferation control by Notch signaling in Drosophila development. Development 125: 2031–2040
Klein T and Arias AM (1999) The vestigial gene product provides a molecular context for the interpretation of signals during the development of the wing in Drosophila. Development 126: 913–925
Maves L and Schubiger G (1998) A molecular basis for transdetermination in Drosophila imaginal discs: interactions between wingless and decapentaplegic signaling. Development 125: 115–124
de Nooij JC, Letendre MA and Hariharan IK (1996) A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87: 1237–1247
Brand AH and Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415
Van de Bor V, Delanoue R, Cossard R and Silber J (1999) Truncated products of the vestigial proliferation gene induce apoptosis. Cell Death Differ. 6: 557–564
Roehm NW, Rodgers GH, Hatfield SM and Glasebrook AL (1991) An improved colorimetric assay for cell proliferation and viability utilizing the tetrazolium salt XTT. J. Immunol. Methods 142: 257–265
Nishimura M, Abiko Y, Mitamura J and Kaku T (1999) Effect of cell plating density and extracellular matrix protein on cell growth of epithelial rests of Malassez in vitro. Med. Electron. Microsc. 32: 127–132
Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M and Dyson NJ (2001) Functional antagonism between E2F family members. Genes Dev. 15: 2146–2160
Yamaguchi M, Hayashi Y and Matsukage A (1995) Essential role of E2F recognition sites in regulation of the proliferating cell nuclear antigen gene promoter during Drosophila development. J. Biol. Chem. 270: 25159–25165
Liu X, Grammont M and Irvine KD (2000) Roles for scalloped and vestigial in regulating cell affinity and interactions between the wing blade and the wing hinge. Dev. Biol. 228: 287–303
Johnston LA and Edgar BA (1998) Wingless and Notch regulate cell-cycle arrest in the developing Drosophila wing. Nature 394: 82–84
Goyal L, McCall K, Agapite J, Hartwieg E and Steller H (2000) Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 19: 589–597
Ryoo HD, Bergmann A, Gonen H, Ciechanover A and Steller H (2002) Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4: 432–438
Gullaud M, Delanoue R and Silber J (2003) A Drosophila model to study the functions of TWIST orthologs in apoptosis and proliferation. Cell Death Differ. 10: 641–651
Nordstrom W, Chen P, Steller H and Abrams JM (1996) Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 180: 213–226
Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM and Abrams JM (2000) Drosophila p53 binds a damage response element at the reaper locus. Cell 101: 103–113
Brennecke J, Hipfner DR, Stark A, Russell RB and Cohen SM (2003) Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36
Bromleigh VC and Freedman LP (2000) p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 14: 2581–2586
Duman-Scheel M, Weng L, Xin S and Du W (2002) Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature 417: 299–304
Hwang MS, Kim YS, Choi NH, Park JH, Oh EJ, Kwon EJ, Yamaguchi M and Yoo MA (2002) The caudal homeodomain protein activates Drosophila E2F gene expression. Nucleic Acids Res. 30: 5029–5035
Kim J, Johnson K, Chen HJ, Carroll S and Laughon A (1997) Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 388: 304–308
Martin-Castellanos C and Edgar BA (2002) A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development 129: 1003–1013
Goux JM and Paillard M (1976) Incomplete penetrance at the vestigal locus of Drosophila. C. R. Hebd. Seances Acad. Sci. D 283: 667–669
Guss KA, Nelson CE, Hudson A, Kraus ME and Carroll SB (2001) Control of a genetic regulatory network by a selector gene. Science 292: 1164–1167
Affolter M and Mann R (2001) Development. Legs, eyes, or wings – selectors and signals make the difference. Science 292: 1080–1081
Baena-Lopez LA and Garcia-Bellido A (2003) Genetic requirements of vestigial in the regulation of Drosophila wing development. Development 130: 197–208
Nagel AC, Wech I and Preiss A (2001) Scalloped and strawberry notch are target genes of Notch signaling in the context of wing margin formation in Drosophila. Mech. Dev. 109: 241–251
Majumdar A, Nagaraj R and Banerjee U (1997) strawberry notch encodes a conserved nuclear protein that functions downstream of Notch and regulates gene expression along the developing wing margin of Drosophila. Genes Dev. 11: 1341–1353
Halder G and Carroll SB (2001) Binding of the Vestigial co-factor switches the DNA-target selectivity of the Scalloped selector protein. Development 128: 3295–3305
Acknowledgements
We are grateful to BA Edgar for helpful comments on the manuscript. We thank SB Carroll, C Lehner, JM Abrams, P Leopold, HD Ryoo, K White and H Steller for fly strains and reagents, and MC Gendron for help with FACS experiments. B Legois and A Skov provided technical assistance, and AM Prêt for comments and corrections with the language. This work was supported by the Association pour la Recherche contre le Cancer (ARC), and an ‘Action Thématique concertée (ATC) vieillissement’ grant from the Institut National pour la Santé Et la Recherche Médicale (INSERM).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Delanoue, R., Legent, K., Godefroy, N. et al. The Drosophila wing differentiation factor Vestigial–Scalloped is required for cell proliferation and cell survival at the dorso-ventral boundary of the wing imaginal disc. Cell Death Differ 11, 110–122 (2004). https://doi.org/10.1038/sj.cdd.4401321
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4401321
Keywords
This article is cited by
-
Wg secreted by conventional Golgi transport diffuses and forms Wg gradient whereas Wg tethered to extracellular vesicles do not diffuse
Cell Death & Differentiation (2021)
-
Exosomal arrow (Arr)/lipoprotein receptor protein 6 (LRP6) in Drosophila melanogaster increases the extracellular level of Sol narae (Sona) in a Wnt-independent manner
Cell Death & Disease (2020)
-
ADAMTS Sol narae cleaves extracellular Wingless to generate a novel active form that regulates cell proliferation in Drosophila
Cell Death & Disease (2019)
-
An ADAMTS Sol narae is required for cell survival in Drosophila
Scientific Reports (2019)
-
From vestigial to vestigial-like: the Drosophila gene that has taken wing
Development Genes and Evolution (2016)