Abstract
A Bcl-2 related family member, Bad, promotes cell death, and its function is regulated by phosphorylation. In this study, we show how the IPNV elicits the induction of Bad gene expression and promotes host apoptotic death. Anti-IPNV-E1S polyclonal and anti-VP3 monoclonal antibodies are used to neutralize the virus that blocks the prime death signal via the virus receptor. In the viability assay, each antibody could also enhance cell viability during IPNV infection. We tested tyrosine kinase inhibitors on IPNV-infected cells in order to assess their effect on blocking the death signal. With 100 μg/ml genistein treatment, Bad-like gene expression was blocked, either by rescuing the IPNV-infected CHSE-214 cells or by blocking internucleosomal DNA cleavage; but the tyrphostin group did not block Bad expression. For CHSE-214 cells, treatment with the protein synthesis-inhibitor, cycloheximide (1 μg/ml), blocked new protein synthesis via activated tyrosine kinase during IPNV infection. We found that Bad protein expression could be blocked, and apoptotic death prevented. Together, these results demonstrate that the IPNV exerts up-regulation of a pro-apoptotic death gene (Bad), the expression of which serves to trigger apoptotic cell death. Our data also suggests that the IPNV induces apoptotic death via a viral receptor which triggers death effector Bad gene expression, possibly through a tyrosine kinase-dependent pathway.
Similar content being viewed by others
Introduction
Apoptosis is the process whereby individual cells of multicellular organisms undergo systematic self-destruction in response to a wide variety of stimuli.1 Apoptosis is a genetically-controlled preprogrammed event which eliminates cells during development when they have become redundant, or, alternatively, a process which functions as an emergency response following radiation damage, viral infection, or aberrant growth of cells as induced by the activation of oncogenes.2
The Bcl-2 family of proteins comprising anti- and pro-apoptotic molecules constitutes a critical, intracellular decision point within a common death pathway.3 The ratio of antagonist (Bcl-2, Bcl-XL, Mcl-1, and A1) to agonist (Bax, Bak, Bcl-XS and Bad) molecules dictates whether a cell will respond to a proximal apoptotic stimulus.3,4 Membership in the family was first defined by homology in two conserved regions, the BH1 and BH2 domains. An additional domain, DH3, has also been noted for Bcl-2 proteins and has proven essential for the pro-apoptotic function of Bak.5,6,7 Bad, was initially identified as a result of its interaction with Bcl-2 and Bcl-XL, and is a distant Bcl-2 family member, bearing only the most universally conserved amino acids within the BH1 and BH2 domains; it lacks the typical hydrophobic C-terminal signal anchor. The presence of Bad counters the anti-apoptotic effect of Bcl-XL and Bcl-2 perhaps by directly inhibiting their activity or by displacing the pro-apoptotic Bax molecule.8 Bad represents a bridging molecule which interconnects signal transduction pathways from extracellular survival factors with the Bcl-2 intracellular checkpoint upon cell death. Bad is phosphorylated on two serine residues embedded in canonical 14-3-3 binding sites in response to the survival factor, IL3.2 Phosphorylated Bad does not bind Bcl-XL and is sequestered in the cytosol, bound to 14-3-3, a specific phosphorylation-binding protein. Substitution at serine phosphorylation sites indicates that phosphorylation of Bad inactivates the molecule and promotes cell survival.9 In more recent studies, Bad has been identified as a potential target of PKB (protein kinase B), linking the PI3K (phosphatidylinositide-3 -OH kinase) pathway directly to the apoptotic machinery.10,11 Bridging of the PI3K-signaling pathway to the Bcl-2 family proteins through Bad represents an attractive model for the pro-survival function of PI3K. The evidence for a connection between PKB and Bad phosphorylation involves expression of a membrane-targeted PKB/Akt, or a kinase-deficient mutant. Cisplatin can induce ovarian cancer cell DNA damage and causes the phosphorylation of both bad Ser-112 via an extracellular signal-regulated protein kinase (ERK) cascade and Bad Ser-136 via a PI-3K-protein kinase B/Akt cascade.12 Insulin can prevent pancreatic β cell apoptotic death through the involvement of sequential induction of IRS-1-associated PI 3-kinase activity, Akt activity, and Bad phosphorylation at Ser-112 and Ser-136.13 On occasion, Bad up-regulation has been reported to lead to cell death as a result of drug treatment.14
The infectious pancreatic necrosis virus (IPNV) is a fish-derived pathogen and is the prototype of the Birnaviridae virus family.15 Birnaviruses possess a bi-segmented, double-stranded RNA genome contained within a medium-sized, unenveloped, icosahedral capsid. Birnavirus gene expression involves the production of four unrelated major genes which undergo various post-translational cleavage processes to generate three to five structural proteins.16 The largest protein, VP1 (90–110 kDa), is encoded by the smaller segment B RNA, and the larger genome segment A encodes VP3 (submajor capsid; 32 kDa), VP4 (28 kDa), and VP2 (major capsid; 46 kDa).17 We are particularly interested in unveiling the molecular mechanisms that regulate the death of cells infected by IPNV. We have previously reported that apoptosis is induced by IPNV infection in a fish cell line18,19 and subsequently found that viral proteins contribute to the down-regulation of the survival factor Mcl-1 during IPNV-induced apoptotic cell death.20 In the present study, we demonstrate that IPNV may induce Bad gene expression through a tyrosine kinase(s)-signaling pathway(s), thus promoting host apoptotic death. This finding may thus provide an important insight into the control of Bad gene expression by viral pathogens.
Results
Visualization of morphological changes of early to late apoptotic CHSE-214 cells
The CHSE-214 cell line was used to test the feasibility of using dual-staining apoptotic cells with ethidium bromide/acridine orange dyes for the detection of apoptotic cells.21 For non-IPNV-infected cells (after 8 h p.i.) as shown in Figure 1A, we found the cells to be labeled with acridine orange only, and the membrane integrity of cells appeared normal. In the early stage, the apoptotic cell was labeled with ethidium bromide/acridine orange dyes revealing an appearance of a rounded cell with the chromatin condensed in the cell (Figure 1B). At this stage (4 h p.i.), the membrane integrity appeared less functional, such that the ethidium bromide could enter the cell, and the condensed chromatin could be labeled with dual dyes at the same time that they appeared orange in color. In the late apoptotic stage (8 h p.i.), the appearances of the rounded cell (long arrow), the post-apoptotic necrosis cell (large short arrow), and apoptotic bodies (small short arrows) are shown by Figure 1C. From Figure 1B,C, we can count the number of viable and apoptotic cells. The phase-contrast image of MOI-1-infected CHSE-214 cells at 12 h p.i. reflecting a rounded cell (long arrow), the formation of membrane blebbing in the apoptotic cell (middle arrow), the appearance of apoptotic bodies (large short arrow), and post-apoptotic necrosis cells (small short arrows) can be seen in Figure 1D. In the apoptotic cell assay, no statistical differences were detected between the apoptotic indices at 0 h (P>0.01) for any samples (Figure 1E). At an MOI of 1, virus infected-CHSE-214 cells at 4 and 8 h p.i. showed higher levels of apoptosis (all P<0.05) than did the negative control (P<0.01). No differences in viability (Figure 1F) were detected among the two groups at 0 h (P<0.01). But the MOI-1 group at 4 and 8 h p.i. showed lower viability (63.4% and 34.1%, respectively; all P<0.05) than did the 1% FBS MEM medium as the negative control (92% and 94%, respectively; all P<0.01).
Phase-contrast fluorescence micrographs of ethidium bromide/acridine orange dye staining of IPNV-infected CHSE cells without infection at 8 h (A) and with MOI 1 at 4 h (B) and 8 h p.i. (C). The apoptotic cells are strongly labeled with dual dyes, cells being labeled with a green and an orange red chromogen. (A) Green fluorescence images of non-infected-CHSE-214 cells, and (B,C) early apoptotic cells (middle arrows), mid apoptotic cell (long arrow) and late apoptotic cells such as apoptotic bodies (small arrows) and post-apoptotic cell (largely show allow) that cells also stained with green, orange and orange red chromogen. (D Phase-contrast micrographs of the morphological characterization of apoptotic IPNV-infected CHSE-214 cells with MOI 1 at 12 h p.i. The different stages of the apoptotic cell reveal the early apoptotic cell (detached and cell round up; indicated by long arrow), middle apoptotic cell (blebbing of cell membrane; middle arrow) and late apoptotic cell including apoptotic bodies (largely short arrows) or the post-apoptotic necrosis cell (small short arrows). (Bar) 10 μm. (E) Apoptosis and cell viability (F) in fish embryo cells as measured by ethidium bromide-acridine staining. The mean of three counts of each different cell (per 200 cells) characteristic was used to calculate the positive apoptotic cells and cell viability indices and their respective bars
Bad is an early response gene during IPNV infection in CHSE-214 cells
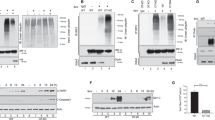
This is the first evidence that the virus can induce expression of the pro-apoptotic gene, Bad. Figure 2A shows that the induction of Bad protein expression by IPNV infection (lane 3) as compared with the negative control (lane 2) has a low expression level. A rat pituitary tumor cell lysate as a positive control is shown in lane 1; it has a similar molecular weight size (23 kDa) to those of lane 1 (human) and lane 3 (fish). The immunoprecipitation method was applied to precipitate the Bad protein from CHSE-214 cells subsequent to IPNV-E1S infection (MOI 1), with the expression patterns and molecular size being assayed by Western blot (Figure 2B). We also found the newly synthesized Bad protein but not the phosphorylated Bad form as shown in lanes 5 to 7 of Figure 2B. The Bad expression pattern in non-infected cells appears in lanes 1 to 4 (Figure 2B). At 6 h p.i., only a small amount of Bad protein can be detected as compared to that found at 0 h p.i. In contrast to this, the IPNV-infected group reveals that the high expression level of Bad protein occurs quickly subsequent to IPNV infection (2 h p.i.; lane 5, Figure 2B) reaching a peak at 4 h p.i. (lane 6, Figure 2B), then slowly decreasing by 6 h p.i. (lane 7, Figure 2B).
Detection of Bad protein expression profile in CHSE-214 by IPNV infection on Western blots. (A) Induction of Bad protein by IPNV infection was examined on Western blots. CHSE-214 cells were infected with IPNV (MOI of 1), and then incubated in MEM containing 1% FCS MEM. Samples were electrophoresed on a SDS-polyacrylamide gel and electro-blotted to a NC membrane. The NC membrane was stained with mouse monoclonal IgG antibodies directed against Bad (Transduction Laboratories). The chemiluminescent signal was imaged on XAR-5 film (Kodak) using a 3-min exposure. Lanes 1, 20 μl of cell lysated from rat pituitary tumor cell line (Transduction Laboratories), Lanes 2, 40 μg of without virus-infected CHSE-214 cells; Lane 3, 40 g of virus-infected (MOI of 1) CHSE-214 cells. (B) Early response gene within apoptotic cell during IPNV infection. CHSE-214 cells were infected with IPNV (MOI of 1), and then incubated in MEM containing 1% FCS MEM. Samples were precipitated with mouse anti-Bad MAb and electrophoresed on a SDS-polyacrylamide gel and electro-blotted to a NC membrane. The NC membrane was stained with mouse monoclonal IgG antibodies directed against Bad (Transduction Laboratories). The chemiluminescent signal was imaged on XAR-5 film (Kodak) using a 5-min exposure. Lanes 1–4, 20 μl of non virus-infected CHSE-214 cells and immuno-precipitated protein corresponding to 0, 2, 4 and 6 h postinfection, respectively; lanes 5–7, 20 μl of virus-infected CHSE-214 cells and immuno-precipitated protein corresponding to 2, 4 and 6 h postinfection, respectively
PolyAb and VP3 MAb used against E1-S virion production prevent cell death
The kind of receptor associated with IPNV capsid proteins VP2 and VP3 during infection has still not been clearly determined. Here, we used the anti-virion of IPNV E1-S polyclonal antibody (1 μg/ml) and the anti-VP3 monoclonal (VP3 8-42-E7 and VP3 8-42-B9) antibody (1 μg/ml) to block E1-S virus infection which may trigger a death signal to activate certain genes. We found that these antibodies (against VP2 and VP3) can also prevent IPNV-induced host-cell death (Figure 3). Especially, anti-E1S whole viral particle polyAb has more effectively prevented cell death than VP3 monoclonal antibodies (8-42-E7 and 8-42-B9) between 16 and 32 h p.i. We found that treatment with either anti-E1-S polyAb (1 μg/ml) or anti-VP3 MAb (8-42-E7) were received more than 100% viability (the time point of 0 h was 100%).
Profiles of effect of anti-E1-S virion polyAb and anti-VP3 MAb as regards the prevention of apoptotic cell death from MOI with 1 of E1-S infected cells at different time points post-infection. At all time points, the number of surviving cells treated with the anti-virion polyAb (1 μg/ml), anti-VP3 MAb (8-42-E7; 1 μg/ml) and 8-42-B9 (1 μg/ml) by IPNV-infected CHSE-214 cells was significantly greater than was the case for those in the non-treated category, either anti-E1-S virion polyAb or anti-VP3 MAb by IPNV-infected CHSE-214 cell. Cell viability at each time point for each cell line was determined in triplicate; each point represents the mean of viability of three independent experiments±S.E.M.
The tyrosine kinase inhibitor, genistein, can block Bad gene expression
In this study, we tried to determine whether any kinase(s) is activated from E1-S associated with the host receptor, with special focus upon the tyrosine kinase that relates to transduction of the death signal for apoptosis during IPNV infection. We tested the ability of the tyrosine kinase inhibitors, genistein (from 1 to 100 μg/ml) and tyrphostin (from 1 to 100 μg/ml), to block Bad gene expression for the potential rescue of IPNV-infected CHSE-214 cells.
In the tyrosine kinase inhibitor assay using the early stress-response gene, Bad, these inhibitors blocked expression when treated with genistein by IPNV infection (Figure 4). The results depicted in Figure 5 reveal that genistein (100 μg/ml) can prevent the morphological changes that occur in apoptotic cells, thus enhancing CHSE-214 cells viability subsequent to IPNV infection at 24 h p.i., although no such observation was made for tyrphostin (from 1 to 100 μg/ml) (data not shown). By contrast, genistein (100 μg/ml) could prevent the intense internucleosomal fragmentation of DNA (Figure 6, lane 6) which was observed for IPNV-infected CHSE-214 cells at 24 h p.i., but such intense internucleosomal fragmentation was not prevented when using genistein at a concentration of either 1 or 10 μg/ml (Figure 6, lanes 4 and 5).
Identification of the effect of blocking Bad protein expression by the tyrosine kinase inhibitor genistein for IPNV-infected CHSE-214 cells using Western blot analysis. After adding tyrosine kinase inhibitor, CHSE-214 cells were infected with IPNV (MOI of 1) and then incubated for one of the following time periods, 0, 2, 4, 6 h, at 18°C. Samples were electrophoresed on a 12% SDS-polyacrylamide gel and electro-blotted to a NC membrane. Antigen-specific signals were detected with a mouse anti-Bad monoclonal antibody. Lane 1, normal CHSE-214 cell lysate; lanes 2–4, IPNV-infected cell lysate for 2, 4 and 6 h post-infection, respectively. Lanes 5–7, depict 30 μg of cell lysate, to which was added genistein (100 μg/ml), followed by IPNV infection and an incubation period for cells the duration of which was 2, 4 and 6 h respectively
The effect of tyrosine kinase inhibitor upon preventing host apoptotic cell death. (A–D) Phase-contrast images illustrating the morphological changes of apoptosis induced in CHSE-214 cell by IPNV infection. (A) Cells in the absence of the viral infection and subsequent to an incubation for 24 h. (B) Cells were infected with MOI of 1 of E1-S for 24 h post-infection. (C) Cells were treated with genistein (100 μg/ml) for 24 h incubation. (D) Cells were treated with genistein (100 μg/ml) then infected with MOI of 1 of E1-S for 24 h post-infection. (Bar) 20 μm. (E) Proportion (%) of effect of genistein (100 μg/ml) on preventing apoptotic cell death from MOI of 1 of E1-S infected cells at different time post-infection. At all time points, the number of surviving cells treated with genistein and IPNV-infected CHSE-214 cells was significantly greater than those for the non-genistein-treated cells and the IPNV-infected CHSE-214 cells. Cell viability at each time point for each cell line was determined in triplicate; each point represents the mean of viability of three independent experiments±S.E.M.
The effect of tyrosine kinase inhibitor upon preventing host cell DNA internucleosomal cleavage prior to IPNV E1-S (MOI of 1) infection. Lane 2: DNA isolated from cells treated with genistein (100 μg/ml), with non IPNV-infected CHSE-214 cells after 24 h postinfection at 18°C used as a negative control. Lane 2: cells infected for 24 h with a MOI of 1 of E1-S. Lanes 4–6: CHSE-214 cells treated with genistein (1, 10 and 100 μg/ml, respectively) in the presence of infection by MOI of 1 of E1-S, respectively, at 24 h post-infection. All samples were electrophoresed through 1.2% agarose gels and visualized by ethidium bromide staining. Lane 1 contains the molecular weight marker used in the gel (1 kb DNA ladder from MBI for sizing linear fragments ranging in size from 500 bp to 1 kb)
Bad protein synthesis can be blocked by the protein synthesis-inhibitor, cycloheximide (CHX)
Interest in apoptosis has prompted a search for genes that encode apoptotic proteins. In the T-cell system, subtractive hybridization studies among T cells suggest that a class of genes that induce RP-8 and PD-1 is transcribed subsequent to the initiating signals for apoptosis,22,23 and if T cells are treated with cycloheximide, the process of apoptosis can be blocked. In our study, we found that the Bad gene can be induced and transcribed after the initiation of an apoptotic signal by IPNV infection. We noted that up to 99% of the production of newly synthesized proteins can be blocked in IPNV-infected CHSE-214 cells during radioactive labeling with [35S]methionine (Figure 7). As indicated by lanes 1 to 7 (Figure 7) which depict the Bad protein expression pattern, the initial expression appears at 2 h p.i., and a major peak follows soon after (6 h p.i.), after which expression was observed to decrease (e.g., as observed at 10 h p.i.), declining further until the low level noted at 24 h p.i.. In addition, for the CHX-treated group, almost 99% of newly synthesized proteins were also blocked including the Bad protein within the time period of from 2 to 24 h p.i. (Figure 7, lanes 8 to 13).
Identification of the effect of protein synthesis-inhibitor blockage upon the newly-synthesized protein. The non-infected cells and the IPNV-infected cells were treated with the protein synthesis inhibitor CHX and were labelled with [35S]methionine at different times and analyzed on 12% SDS–PAGE. Lane 1, [35S]methionine-labelled CHSE-214 cell lysed after 2 h; lanes 2–7, labelled newly-synthesized polypeptide release by virus-infected cells lysed after 2, 4, 6, 8, 10 and 24 h post-infection, respectively; lanes 8–13, IPNV-infected cells treated with CHX (1 μg/ml) and lysed following 2, 4, 6, 8, 10 and 24 h of CHX treatment, respectively
Also evident from Figure 7 is that the Bad protein can be blocked as described above; we noted that such inhibition can prevent the morphological changes attributable to IPNV infection of CHSE-214 cells (Figure 8A–H) and can also enhance these cells viability (Figure 8I). Finally, as revealed by Figure 9, intense internucleosomal DNA cleavage occurring between 6 and 8 h p.i. (lanes 5 and 6; Figure 9) was consistently observed, which corresponds to a major Bad expression peak (Figure 7, lane 7) that was effectively blocked in the CHX-treated group (lanes 8 to 13).
The effect of protein synthesis inhibitor upon the prevention of host apoptotic cell death. (A–H) Phase-contrast images illustrating the morphological changes of apoptosis induced for CHSE-214 cells by IPNV infection. (A–C) Cells incubated in the absence of the viral infection for 6, 12 and 24 h, respectively. (D) Cells treated with CHX (1 μg/ml) for 24 h incubation. (E–G) Cells infected with MOI of 1 of E1-S for 6, 12 and 24 h post-infection, respectively. (H) Cells treated with CHX (1 μg/ml) and with MOI of 1 of E1-S infection for 24 h post-infection. (Bar) 20 μm. (I) Percentage of effect of CHX on preventing apoptotic cell death from both non- and MOI of 1 of E1-S infection group at different time points post infection. At all time points, the number of surviving cells treated with CHX and IPNV-infected CHSE-214 cells was significantly greater than those in the non-CHX treated IPNV-infected CHSE-214 cells. Cell viability at each time point for each cell line was determined in triplicate; each point represents the mean of viability of three independent experiments±S.E.M.
The effect of protein synthesis inhibitor upon preventing host cell DNA internucleosomal cleavage prior to IPNV E1-S (MOI of 1) infection. Lanes 2–7: DNA isolated from IPNV-infected (MOI of 1) CHSE-214 cells following periods of 0, 2, 4, 6, 8 and 10 h post-infection, respectively, at 18°C. Lane 8: cells were treated with CHX (1 μg/ml), without infection for an incubation period of 10 h. Lanes 9–13: cells were treated with cHX (1 μg/ml) then infected by a MOI of E1-S for 2, 4, 6, 8 and 10 h post infection, respectively. All samples were electrophoresed through 1.2% agarose gels and visualized by ethidium bromide staining. Lane 1 contains the molecular weight marker used in the gel (1 kb DNA ladder from MBI for sizing linear fragments ranging in size from 500 bp to 1 kb)
Discussion
Infectious pancreatic necrosis virus (IPNV) is a fish pathogen. E1-S of the IPNV Ab strain induces apoptotic cell death in CHSE-214 cells as suggested previously by Hong18 and induces apoptotic death in zebrafish ZFL cells (data not shown). In this study, we examined the newly emergent virus from double-stranded RNA virus-IPNV and attempted to interpret how the virus triggers a death signal for the transcription and translation of a Bad death effector gene leading to the induction of host apoptotic cell death.
Bad belongs to a pro-apoptotic subfamily that can be initially identified by its interaction with Bcl-2 and Bcl-XL. Bad represents a bridging molecule interconnecting a signal transduction pathway between extracellular survival factors and the Bcl-2 intracellular checkpoint upon cell death. Bad is phosphorylated at two serine residues embedded in canonical 14-3-3 binding sites in response to a survival factor, IL3.2 The occurrence of this substitution at the serine phosphorylation sites indicates that phosphorylation of Bad has inactivated the molecule to promote cell survival;9 it behaves as a potential target of PKB, linking the PI3K pathway directly to the apoptotic machinery.10,11 Most recently, 14-3-3 proteins and survival kinases have been reported to be able to cooperate in order to inactivate Bad by BH3-domain phosphorylation.24 In our study, we found that Bad expression may act as an early stress response gene during IPNV infection (as shown in Figure 2) which differs markedly from current opinion (as described above). We are very interested in examining the point of death signal triggering which induces Bad gene expression in our cell model system.
We do know that the best-understood initiating events of apoptosis are those involving the cell surface transmembrane tumor necrosis factor (TNF)-receptor (TNFR) superfamily of death receptors,25 although we remain uncertain regarding the role of a specific IPNV-infected receptor. We used the anti-virion polyAb (demonstrating activity against major capsid, VP2, and submajor capsid protein, VP3 polyAb) and anti-submajor capsid protein, VP3 MAb, in order to neutralize the virion that could either block the virion associated with infected-specific receptor or prevent its entry into the cell for replication. Further, we also found that host-cell apoptotic death can be blocked (Figure 3), revealing that the viral receptor is strongly related to the process of host-cell apoptosis induction.
The question remains: What death signal is triggered by IPNV to induce CHSE-214 apoptosis? Apoptosis is a highly regulated process of cell death. As for cell proliferation and differentiation, apoptosis is controlled by hormonal and other receptor-mediated cues. Receptor-mediated cellular control mechanisms, in general, act through a limited set of signal transduction systems that release secondary messenger molecules and elicit the activation of a protein kinase/phosphatase cascade, by altering the phosphorylation status of key target proteins which regulate cellular metabolism and gene expression.26 For thymocytes, the anti-CD3 MAb-induced thymic apoptosis process depends upon protein tyrosine kinase (PTK) activation via TCR, and the tyrosine kinase inhibitors, genistein or herbimycin, can prevent apoptotic death.27 In our study, we found that some cellular proteins can be phosphorylated at tyrosine residues (data not shown) during IPNV infection, and the tyrosine kinase inhibitor, genistein (100 μg/ml), can also block Bad gene expression (Figure 4), and prevent both the apoptotic morphological changes which occur in IPNV infected-CHSE-214 cells and the formation of internucleosomal DNA fragments (Figure 5), although we did not make similar observations for tyrphostin.28
In the meantime, we even tested the growth factors including human IGF-I, IGF-II, and growth hormone to see whether they can prevent apoptotic death from E1-S infection of host cells. With the tested dosage of from 1 ng/ml to 1 μg/ml, we found that none could prevent host death during IPNV infection (data not shown). We propose that there may be a very different regulation system from human insulin which enhances pancreatic β cell survive through PI-3-kinase activity and Akt phosphorylation of Bad at serine 112 and 136.21
Thus, we were interested to know whether Bad expression is essential for inducing the CHSE-214 apoptotic process or not. Apoptotic cells death is perceived as a type of cell suicide, since it appears to be genetically controlled. This view stems from studies which suggest that transcriptional and/or translational control over the apoptotic process is maintained by the dying cell. For example, some processes, including apoptosis of rodent thymocytes induced by treatment with the Ca2+ ionophore, A23187,29 the action of certain cytotoxic drugs or nucleosides,30,31 the apoptosis of T-cell lines induced by IL-2 withdrawal,32 and the apoptosis of T-cell hybridomas induced by anti-CD3 antibody treatment,33 all relate to a delay in or an abrogation of inhibitors of RNA or protein synthesis in every case. Such studies have led to the notion that macromolecular synthesis is an essential component of the apoptotic response in general, and that this represents the synthesis of cell suicide-programmed components that are encoded by a putative death gene. In our study, we found that the putative death gene, the Bad gene, is quickly induced and expressed at 2 h p.i. with IPNV infection (Figures 2 and 4). The major peak in Bad expression (Figure 7, lane 4) may correlate with the induction of a DNA fragment between 6 and 8 h p.i. (Figure 9, lanes 5 and 6). During the blockage of Bad gene expression by CHX treatment (Figure 7, lanes 8 to 13), features of apoptosis such as associated degenerative morphological changes, DNA fragmentation, and decreasing cell viability are blocked and cell survival enhanced (Figures 8 and 9). It would appear, however, that Bad gene expression is essential for inducing CHSE-214 apoptosis (Figures 7,8,9), although much still remains unclear as to whether the Bad gene regulates other known death effectors.
We know that all viruses infect specific target cells, replicating large numbers of progeny virions, and spreading these progeny to initiate new rounds of infection. Viruses encode proteins which are highly efficient for the optimization of such replication; however, target organisms use both systemic and cell-based defense mechanisms to limit the extent of viral infection, including immune and inflammatory processes and the execution or suicide of infected cells. Interestingly, in our study, we found that IPNV exerts a viral gene, VP5, that is a novel anti-apoptosis member of the Bcl-2 family, in order to up-regulate the host survival factor Mcl-1 to enhance viral progeny production (submitted for publication). This phenomenon may explain how convergent evolution ablates the host response by targeting strategic points in specific defense pathways that are able to inhibit or delay protective actions until sufficient viral yields have been produced.33,34,35 In our study, however, we examined the process of IPNV-induced apoptosis triggering a death signal from the viral receptor which is subsequently transcribed and translated to the Bad death gene through a protein tyrosine kinase-dependent pathway, and we summarize these findings and combine the activity of the anti-apoptotic gene, VP5, in order to demonstrate the process of regulating the cell-death pathway (Figure 10).
The apoptotic cascade. The cell is exposed to a stimulus that is transmitted through the cellular machinery during the signaling phase. For instance, when a cell is infected with IPNV-E1-S, the virus associated with the cellular receptor triggers a death-signalling cascade that may be activate tyrosine kinase, and then induce the Bad death gene expression at the effector stage. In the degradation stage, phosphatidylserine (PS) and endonuclease is externalized and activates that cells cytoskeleton, and DNA is restructured and undergoes cleavage before the cell body finally enters secondary apoptotic necrosis. Death-receptor signaling is halted by anti-capsid protein antibodies interfering with (a) a virus-receptor interaction, specific tyrosine kinase inhibitor genistein (b) block-specific tyrosine kinase activity, the protein synthesis-inhibitor cycloheximide (CHX) (c) block Bad protein translation processes and VP5, a novel anti-apoptotic Bcl-2 family member (d) block nuclease activity
Materials and Methods
Cells and virus
Chinook salmon embryo cells (CHSE-214) were obtained from the American Type Culture Collection (ATCC). These CHSE-214 cells were grown at 18°C in plastic tissue-culture flasks (Nunc) using Eagle's minimum essential medium (MEM) supplemented with 10% (v/v) fetal calf serum and 25 μg/ml of gentamicin. The isolated virus, E1-S, a member of the Ab strain of IPNV, was obtained from Japanese eels in Taiwan.36 The E1S virus was propagated in CHSE-214 cell monolayers at a 0.01 multiplicity of infection (MOI) per cell. Infected cultures were incubated at 18°C until an extensive cytopathogenic effect (CPE) was observed.37 Virus plaque assays38 and TCID50 were performed on confluent monolayers of CHSE-214 cells.36
Double staining apoptotic cells by ethidium bromide/acridine orange dyes
Cells (103 cells/0.1 ml culture medium) were seeded into two wells of a chamber slide (Nunc) and incubated at 18°C for 20 h. Prior to infection, the 10% FBS MEM medium used for cell culture medium was changed to 1% FBS MEM. At 4 and 8 h subsequent to this medium change, a sample from each group was removed from the medium and washed with PBS (pH 7.4), and then stained with 1 μl of acridine orange (100 μg/ml)-ethidium bromide (100 μg/ml) in 0.5 ml PBS for from 2 to 5 min at room temperature. The stained cells were placed on a glass slide and covered with a 22-mm2 coverslip. Slides were examined by fluorescent microscopy using an Olympus IX70 microscope equipped with a BP450-480 band-pass excitation filter and a BA515 barrier-emission filter for observation of dual dye-treated samples. Each group sample (two wells) was counted three times, and each time, 200 or more cells were counted. The characteristics of cells were recorded according to the color and structure of the chromatin. The mean of the three counts of each different cell characteristic was used to calculate the apoptotic and cell viability indices and their respective bars.
Production of polyclonal antibody (polyAb) and monoclonal antibodies (MAb) against the IPNV-E1S virion and VP3 submajor capsid protein
A New Zealand rabbit was subcutaneously immunized twice every 2 weeks with 500 μg/1.2 ml of purified IPNV-E1S virion emulsified in an equal volume of Freund's complete adjuvant, boosted with 600 μg of antigen emulsified in an incomplete adjuvant at the seventh week. The rabbit was bled at the end of the eighth week, and 30 ml was removed. Collected serum was precipitated with 50% (NH4)2SO4, dialyzed against a 0.1 M phosphate buffer pH 8.0 (PB), and purified by protein A chromatography (Pharmacia, Sweden). The immunoglobulin fraction eluted at pH 3.0 was collected and dialyzed against phosphate buffered saline (PBS). The resultant anti-E1-S polyclonal antibodies were aliquoted and frozen at −70°C.
Three BALB/c mice were each subcutaneously immunized with four bi-weekly infections, comprising 50 μg each of purified IPNV-E1S virion emulsified in 0.2 ml Freund's complete adjuvant. Every mouse was intravenously boosted with 50 μg of IPNV-E1S virion 3 days before fusion. Splenic lymphocytes from immunized mice were fused to BALB/c myeloma FO obtained from the American Type Cell Culture (ATCC) laboratories. Hybridomas were selected in HAT-10% fetal calf serum (FCS)-RPMI medium as previously described by Hong et al.39
Quantification of cell viability
About 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultured for more than 20 h. To test if antibodies could prevent IPNV E1-S from inducing cell death, confluent cells in 60-mm-diameter plastic tissue culture plates (Nunc) were pretreated with either 1 μg/ml anti-virion polyAb of E1-S or 1 μg/ml anti-Mab of VP3 (8-42-E7 and 8-42-B9) for 2 h, and then these infected-CHSE-214 cells (MOI 1) were incubated for one of the following time periods: 0, 16, or 32 h. For assayed groups using genistein (100 μg/ml) or cycloheximide (1 μg/ml) (CHX) to prevent cell death, confluent cells were cultured in 60-mm-diameter plastic tissue culture plates (Nunc) that had been treated with 100 μg/ml genistein and 1 μg/ml CHX, then infected with MOI 1 of IPNV E1-S for one of the following time periods: 0, 6, 12, or 24 h in 1% FBS MEM at 18°C. At the completion of the various culture periods, cell layers contained in the culture vessels were washed with PBS, and the monolayers were treated with 0.5 ml of 0.1% trypsin-EDTA (GIBCO) for from 1 to 2 min. Cell viability was determined using a trypan blue dye exclusion assay.40 Cell viability at each time point for each cell line was determined in triplicate; each point represents the mean of viability of three independent experiments±the standard error of the mean (S.E.M.). Data were analyzed using either paired or unpaired Student's t-test as appropriate. A value of P<0.05 was taken to represent a statistically significant difference between mean values of groups.
Immunoprecipitation of the Bad protein
In the Bad protein expression assay, about 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultured for more than 20 h. The cell monolayers so produced were rinsed twice with PBS; the medium was then changed to 1% FBS MEM, and the cells were infected with an MOI of 1 of IPNV and incubated for 4 h. Uninfected control cells were also incubated for the same periods of time. At the end of the incubation period, the culture medium was aspirated, and cells were washed with PBS, following which they were lysed in 0.3 ml of lysis buffer (10 mM Tris-base, 20% glycerol, 10 mM SDS, 2% β-ME, pH 6.8).
Proteins present in the cell lysate were separated by SDS-polyacrylamide gel electrophoresis,41 electro-blotted, and subjected to immunodetection as described by Kain et al.42 In the immunoprecipitation of Bad protein expression assay, about 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultured for more than 20 h. The cell monolayers so produced were rinsed twice with PBS; the medium was then changed to 1% FBS MEM, and cells were infected with an MOI of 1 of IPNV and incubated for one of the following time periods: 0, 2, 4, or 6 h. Uninfected control cells were also incubated for the same periods of time. At the end of each incubation period, the culture medium was aspirated, and cells were washed with PBS, following which they were lysed in 1 ml of lysis buffer (0.5% (v/v) Nonidet P-40 in phosphate-buffered saline). Cell extracts were incubated for 18 h at 4°C in the presence of 0.1% (w/v) N-laurolysarcosine, 1% bovine serum albumin, 10 M phenylmethanesulfonyl fluoride (PMSF), and anti-mouse Bad monoclonal antibodies (Transduction Laboratories) at a dilution of 1 : 500. Following this, 160 μl of 10% (v/v) protein A-Sepharose (Zyme) in 150 mM NaCl/10 mM Tris-HCl, pH 7.8/1% N-lauroylsarcosine was added, and the mixture was incubated at 4°C for 1 h. The protein A-Sepharose-antibody complexes so formed were washed three times in the same buffer and boiled in 200 μl of 10 mM Tris-HCl, pH 7.8/1 mM EDTA/0.1% SDS.43 Bound proteins were released by boiling the beads in 30 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer containing 5% SDS and 4% β-mercaptoethanol.37 Proteins present in the cell lysate were separated by SDS-polyacrylamide gel electrophoresis,41 electro-blotted, and subjected to immunodetection as described by Kain et al.42
Immunoblotting
Approximately 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultivated for more than 20 h. Cell monolayers were rinsed twice with PBS, following which the medium was changed to 1% FBS MEM treated with 100 μg/ml of the tyrosine kinase-inhibitor, genistein. These cells were then infected with an MOI of 1 of IPNV and incubated for one of the following time periods: 0, 2, 4, or 6 h. Uninfected control cells were also treated with 100 μg/ml genistein and incubated for analogous periods of time. At the completion of each incubation period, the culture medium was aspirated, and cells were washed with PBS and then lysed in 0.3 ml lysis buffer (10 mM Tris-base, 20% glycerol, 10 mM SDS, 2% β-ME, pH 6.8).
Proteins present in the cell lysate were separated by SDS-polyacrylamide gel electrophoresis,41 electro-blotted, and subjected to immunodetection as described by Kain et al.42 Blots were incubated with a 1 : 500 dilution of anti-mouse Bad monoclonal antibody and a 1 : 7500 dilution of peroxidase-labeled goat anti-rabbit conjugate (Amersham). Chemiluminescent detection was performed according to the instructions provided with the Western Exposure Chemiluminescent Detection System (Amersham). Chemiluminescent signals were imaged by exposure of Kodak XAR-5 film (Eastman Kodak, Rochester, NY, USA).
DNA preparation and gel electrophoresis
For the genistein experiment, approximately 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultivated for more than 20 h. CHSE-214 cell monolayers were then changed to 1% FBS MEM and treated with 1, 10, 100 μg/ml of genistein. Cells were then infected with an MOI of 1 for 24 h. Uninfected control cells were treated with 100 μg/ml genistein and incubated for 24 h. For the cycloheximide (CHX) experiment, approximately 105 CHSE-214 cells/ml were seeded in a 60-mm petri dish and cultivated for more than 20 h. The resultant CHSE-214 cell monolayers were then incubated in 1% FBS MEM either in the presence or absence of CHX at 1 μg/ml. Cells were then infected with an MOI of 1 for one of the following time periods: 0, 2, 4, 6, 8, or 10 h. Uninfected control cells were treated either in the presence or the absence of CHX (1 μg/ml) for either 0 or 10 h. Following the completion of incubation, cells were lysed with lysis buffer (10 mM Tris-HCl, 0.25% Triton X-100, 1 mM EDTA, pH 7.4). Following treatment of the cell lysate with phenol-chloroform-isoamyl alcohol (25 : 24 : 1), genomic DNA was precipitated in the presence of 0.3 M sodium acetate and cold absolute ethanol at −70°C for 2 h and then resuspended in 10 mM Tris-HCl (pH 7.4) and 1 mM EDTA. Aliquots of 20 μl of this suspension containing approximately 5 to 10 μg of genomic DNA were then electrophoresed in 1.2% agarose gels for a period of 2 h at 40 V. Gels were subsequently stained with ethidium bromide and photographed under UV transillumination.18
Radioactive labeling of infected cells for polyacrylamide gel electrophoresis
Confluent CHSE-214 cell monolayers were cultivated in 60-mm-diameter plastic tissue culture plates (Nunc) for more than 20 h. The resultant cell monolayers were rinsed twice with PBS, following which starvation of cells in serum-free medium (Gibco) was conducted for 2 h. Cells were subsequently treated, either in the presence or absence of CHX at 1 μg/ml, with an MOI of 1 of IPNV as an infecting agent, following which 50 μCi/ml of [35S]methionine at 18°C was added for one of the following time periods: 2, 4, 6, 8 or 10 h. At completion of the labeling period, the cell layer was washed with PBS, and the monolayer was lysed in 0.4 ml of electrophoresis sample buffer, and heated for 2 min in a boiling water bath. Radioactively-labeled proteins were analyzed in the slab gels using the discontinuous SDS gel system described by Laemmli.41 The 12% separating gel and 5% stacking gel were prepared at a ratio of acrylamide to bisacrylamide selected to be 30 : 0.8. Sixty microliters of labeled cell lysate in a total volume of 400 μl was loaded into each well. Electrophoresis was conducted for 14 to 16 h at 60 V. The slab gel was dried under a vacuum, then autoradiography was performed and the exposed film developed.
Abbreviations
- CHX:
-
cycloheximide
- CPE:
-
cytopathogenic effect
- IPNV:
-
infectious pancreatic necrosis virus
- MAb:
-
monoclonal antibody
- MOI:
-
multiplicity of infection
- NSP:
-
newly-synthesized protein
- PI3K:
-
phosphatidylinositide
- PKB:
-
protein kinase B
- PolyAb:
-
polyclonal antibody
- PTK:
-
protein tyrosine kinase
- TNFR:
-
tumor necrosis factor-receptor
- VP:
-
viral protein
References
Wyllie AH, Kerr JFR, Currie AR . 1980 Cell death: the significance of apoptosis Int. Rev. Cytol. 68: 251–306
Newton K, Strasser A . 1998 The Bcl-2 family and cell death regulation Curr. Opin. Genet. Dev. 8: 68–75
Farrow SN, Brown R . 1996 New members of the BCL-2 family and their protein partners Curr. Opin. Genet. Dev. 6: 45–49
Oltvai ZN, Milliman CL, Korsmeyer SJ . 1993 Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death Cell 74: 609–619
Chittenden T, Harrington EA, O'Connor R, Flemington C, Lutz RJ, Evan G, Guild BC . 1995 Induction of apoptosis by the Bcl-2 homologue Bak Nature 374: 733–736
Hunter JJ, Parslow TG . 1996 A peptide sequence from Bax that converts Bcl-2 into an activator of apoptosis J. Biol. Chem. 271: 8521–8524
Zha H, Aime-Sempe C, Sato T, Reed JC . 1996 Propoptotic protein Bax heterodimerizes with Bcl-2 and homodimerizes with Bax via a novel domain (BH3) distinct from BH1 and BH2 J. Biol. Chem. 271: 7440–7444
Yang E, Zha J, Jokel J, Boise LH, Thompson CB, Korsmeyer SJ . 1995 Bad, a heterodimeric partner for Bcl-xL and Bcl-2, displaces Bax and promotes cell death Cell 80: 285–291
Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ . 1996 Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X Cell 87: 619–628
Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME . 1997 Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery Cell 91: 231–241
del Peso L, Gonzalez-Garcia M, Page C, Herrfera R, Nunez G . 1997 Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt Science 278: 687–689
Hayakawa J, Ohmichi M, Kurachi H, Kanda Y, Hisamoto K, Nishio Y, Adachi K, Tasaka K, Kanzaki T, Murata Y . 2000 Inhibition of Bad phosphorylation either at serine 112 via extracellular signal-regulated protein kinase cascade or at serine 136 via Akt cascade sensitizes human ovarian cancer cells to cisplatin Cancer Res. 60: 5988–5994
Federici M, Hribal ML, Ranalli M, Marselli L, Porzio O, Melino G, Sesti G . 2001 The common Arg972 polymorphism in insulin receptor substrate-1 causes apoptosis of human pancreatic islets FASEB J. 15: 22–24
Tudor G, Aguilera A, Halverson HO, Laing ND, Sausville EA . 2000 Susceptibility to drug-induced apoptosis correlates with differential modulation of Bad, Bcl-2 and Bcl-xL protein levels Cell Death Differ. 7: 574–586
Dobos P, Hill BJ, Hallett R, Kells DTC, Becht H, Tenings D . 1979 Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes J. Virol. 32: 593–605
Dobos P . 1995 The molecular biology of infectious pancreatic necrosis virus (IPNV) Annu. Rev. Fish Dis. 5: 25–54
Wu JL, Hong JR, Chang CY, Hui CF, Liao CF, Hsu YL . 1998 Involvement of serine proteinase in infectious pancreatic necrosis virus capsid protein maturation and NS proteinase cleavage in CHSE-214 cells J. Fish Dis. 21: 215–220
Hong JR, Lin TL, Hsu YL, Wu JL . 1998 Apoptosis procedes necrosis of fish cell line by infectious pancreatic necrosis virus Virology 250: 76–84
Hong JR, Lin TL, Yang JY, Hsu YL, Wu JL . 1999a Dynamics of nontypical apoptotic morphological changes visualized by green fluorescent protein in living cells with infectious pancreatic necrosis virus infection J. Virol. 73: 5056–5063
Hong JR, Hsu YL, Wu JL . 1999b Infectious pancreatic necrosis virus induces apoptosis due to down-regulation of survival factor MCL-1 protein expression in a fish cell line Virus Res. 63: 75–83
Duke RC, Cohen JJ . 1992 Morphological and biochemical assays of apoptosis In Current Protocols in immunology Vol. 1: ed. Coligan J, Kruisbeek AM, Argulies DH, Evach EM, Strober W John Wiley and Sons, New York, NY pp 3.17.1–3.17.16
Cohen JJ, Duke RC, Fadok VA, Sellins KS . 1992 Apoptosis and programmed cell death in immunity Annu. Rev. Immunol. 10: 267–293
Ishid Y, Agata Y, Shibahara K, Honjo T . 1992 Induced expression of the immunoglobulin gene superfamily, upon programmed cell death EMBO J 11: 3887–3895
Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME . 2000 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation Mol. Cell. 6: 41–51
Ashkenazi A, Dixit VM . 1998 Death receptors: signaling and modulation Science 281: 1305–1308
McConkey D, Orrenius S . 1994 Signal transduction pathways to apoptosis Trends. Cell Biol. 4: 370–374
Migita K, Eguchi K, Kawabe Y, Mizokami A, Tsukada T, Nagataki S . 1994 Prevention of anti-CD3 monoclonal antibody-induced thymic apoptosis by protein tyrosine kinase inhibitors J. Immunol. 153: 3457–3465
Lyall RM, Zilberstein A, Gazit A, Gilon C, Levitzki A, Schlessinger J . 1989 Tyrphostins inhibit epidermal growth factor (EGF)-receptor tyrosine kinase activity in living cells and EGF-stimulated cell proliferation J. Biol. Chem. 264: 14503–14509
Kizaaki H, Tadakuma T, Odaka C, Muramatsu J, Ishimura Y . 1989 Tumor necrosis factor-alpha enhances cAMP-induced programmed cell death in mouse thymocytes J. Immunol. 143: 1790–1794
Kizaaki H, Shimada H, Ohsaka F, Sakurada T . 1988 Adenosine, deoxyadenosine, and deoxyguanosine induce DNA cleavage in mouse thymocytes J. Immunol. 141: 1652–1657
McConkey DJ, Hartzell P, Nicotera P, Wyllie AH, Orrenius S . 1988 Toxicol. Lett. 42: 123–130
Cohen JJ, Duke RC, Chervenak R, Sellins KS, Olson LK . 1985 DNA fragmentation in targets of CTL: an example of programmed cell death in the immune system Adv. Exp. Med. Biol. 184: 493–506
Shi Y, Szalay MG, Paskar L, Boyer M, Singh B, Green DR . 1990 Activation-induced cell death in T -cell hybridomas is due to apoptosis morphological aspects and DNA fragmentation J. Immunol. 144: 3326–3333
Tschopp J, Thome M, Hofmann K, Meinl E . 1998 The fight of viruses against apoptosis Curr. Opin. Genet. Dev. 8: 82–87
Roulston A, Marcellus RC, Rranton PE . 1999 Viruses and apoptosis Annu. Rev. Microbiol. 53: 577–628
Wu JL, Chang CY, Hsu YL . 1987 Characteristics of an infectious pancreatic necrosis-like virus isolated from Japanese eel (Anguilla japonica) Bull. Inst. Zool., Academia Sinica 26: 201–214
Dobos P . 1977 Virus-specific protein synthesis in cells infected by infectious pancreatic necrosis virus J. Virol. 21: 242–258
Nicholson B, Dunn J . 1974 Homologous viral interference in trout and Atlantic salmon cell cultures infected with infectious pancreatic necrosis virus J. Virol. 14: 180–182
Hong TH, Chen ST, Tang TK . 1989 The production of polyclonal and monoclonal antibodies in mice using novel immunization methods J. Immunol. Methods 120: 151–157
Mullen OL, Dodd MC, Minton JP . 1975 Evaluation of dye exclusion and colony inhibition techniques for detection of polyoma-specific, cell-mediated immunity J. Natl. Cancer Inst. 54: 229–231
Laemmli UK . 1970 Cleavage of structural proteins during the assembly of the head of bacteriophage T4 Nature 227: 680–685
Kain SR, Mai K, Sinai P . 1994 Human multiple tissue Western blots: a new immunological tool for the analysis of tissue-specific protein expression BioTechniques 17: 982–987
Hanecak K, Semler BL, Anderson CW, Wimmer E . 1982 Proteolytic processing of poliovirus polypeptides: Antibodies to polypeptide p3-7c inhibit cleavage at glutamine-glycine pairs Proc. Natl. Acad. Sci. USA 79: 3973–3977
Acknowledgements
This work was supported by grants (NSC-89-2311-B-001-005, NSC-90-2311-B-001-018) awarded to Dr. Jen-Leih Wu from the National Science Council, Taiwan, Republic of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by M Piacentini
Rights and permissions
About this article
Cite this article
Hong, JR., Wu, JL. Induction of apoptotic death in cells via Bad gene expression by infectious pancreatic necrosis virus infection. Cell Death Differ 9, 113–124 (2002). https://doi.org/10.1038/sj.cdd.4400933
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.cdd.4400933
Keywords
This article is cited by
-
Interplay of cell death signaling pathways mediated by alternating magnetic field gradient
Cell Death Discovery (2018)
-
Knockdown of zebrafish YY1a can downregulate the phosphatidylserine (PS) receptor expression, leading to induce the abnormal brain and heart development
Journal of Biomedical Science (2016)
-
GSIV serine/threonine kinase can induce apoptotic cell death via p53 and pro-apoptotic gene Bax upregulation in fish cells
Apoptosis (2016)
-
ORF005L from infectious spleen and kidney necrosis virus is located in the inner mitochondrial membrane and induces apoptosis
Virus Genes (2014)
-
Beta-nodavirus B2 protein induces hydrogen peroxide production, leading to Drp1-recruited mitochondrial fragmentation and cell death via mitochondrial targeting
Apoptosis (2014)