Abstract
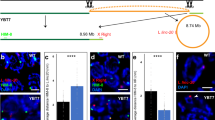
Sex is determined in Caenorhabditis elegans by an X-chromosome-counting mechanism that reliably distinguishes the twofold difference in X-chromosome dose between males (1X) and hermaphrodites (2X)1,2. This small quantitative difference is translated into the ‘;on/off’ response of the target gene, xol-1, a switch that specifies the male fate when active and the hermaphrodite fate when inactive3. Specific regions of X contain counted signal elements whose combined dose sets the activity of xol-1 (ref. 4). Here we ascribe the dose effects of one region to a discrete, protein-encoding gene, fox-1. We demonstrate that the dose-sensitive signal elements on chromosome X control xol-1 through two different molecular mechanisms. One involves the transcriptional repression of xol-1 in XX animals. The other uses the putative RNA-binding protein encoded by fox-1 to reduce the level of xol-1 protein. These two mechanisms of repression act together to ensure the fidelity of the X-chromosome counting process.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Nigon, V. Polyploidie experimentale chez un nematode libre, Rhabditis elegans Maupas. Bull. Biol. Fr. Belg. 85, 187–225 (1951).
Madl, J. E. & Herman, R. K. Polyploids and sex determination in Caenorhabditis elegans. Genetics 93, 393–402 (1979).
Rhind, N. R., Miller, L. M., Kopczynski, J. B. & Meyer, B. J. xol-1 acts as an early switch in the C. elegans male/hermaphrodite decision. Cell 80, 71–82 (1995).
Akerib, C. C. & Meyer, B. J. Identification of X chromosome regions in Caenorhabditis elegans that contain sex-determination signal elements. Genetics 138, 1105–1125 (1994).
Bridges, C. B. Non-disjunction as proof of the chromosome theory of heredity. Genetics 1, 1–52 (1916).
Cline, T. W. & Meyer, B. J. Vive la différence: males vs females in flies vs worms. Annu. Rev. Genet. 30, 637–702 (1996).
Miller, L. M., Plenefisch, J. D., Casson, L. P. & Meyer, B. J. xol-1: A gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55, 167–183 (1988).
Meyer, B. J. & Casson, L. P. Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47, 871–881 (1986).
Hodgkin, J., Zellan, J. D. & Albertson, D. G. Identification of a candidate primary sex determination locus, fox-1 on the X chromosome of Caenorhabditis elegans. Development 120, 3681–3689 (1994).
Bird, C. G. & Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 265, 615–621 (1994).
Willard, H. F. Xchromosome inactivation, XIST, and pursuit of the X-inactivation center. Cell 86, 5–7 (1996).
Barstead, R. J., Kleiman, L. & Waterston, R. H. Cloning, sequencing and mapping of an α-actinin gene from the nematode Caenorhabditis elegans. Cell Motil. Cytoskel. 20, 69–78 (1991).
Fire, A., Harrison, S. W. & Dixon, D. Amodular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93, 189–198 (1990).
Hodgkin, J., Horvitz, H. R. & Brenner, S. Nondisjunction mutants of Caenorhabiditis elegans. Genetics 91, 67–94 (1979).
Chuang, P.-T., Albertson, D. G. & Meyer, B. J. DPY-27: A chromosome condensation homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79, 459–474 (1994).
Acknowledgements
We thank I. Carmi, T. Cline, J. Rine, H. Dawes for discussions and critical comments on the manuscript, A. Fire for reporter gene vectors and Y. Jin for the gfp coding sequences. M.N. is a National Science Foundation predoctoral fellow. C.C.A. was an American Cancer Society postdoctoral fellow. This work was supported by a grant from the National Institutes of Health to B.J.M.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nicoll, M., Akerib, C. & Meyer, B. X-chromosome-counting mechanisms that determine nematode sex. Nature 388, 200–204 (1997). https://doi.org/10.1038/40669
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/40669
This article is cited by
-
Evolution of sexual systems, sex chromosomes and sex-linked gene transcription in flatworms and roundworms
Nature Communications (2022)
-
Dosage compensation and its roles in evolution of sex chromosomes and phenotypic dimorphism: lessons from Drosophila, C.elegans and mammals
The Nucleus (2017)
-
Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans
Nature Cell Biology (2009)
-
Fox-1 family of RNA-binding proteins
Cellular and Molecular Life Sciences (2009)
-
The worm solution: a chromosome-full of condensin helps gene expression go down
Chromosome Research (2009)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.