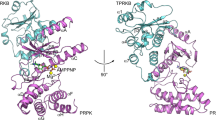
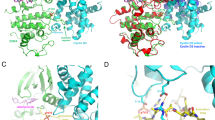
Abstract
In cancer, the biochemical pathways that are dominated by the two tumour-suppressor proteins, p53 and Rb, are the most frequently disrupted. Cyclin D-dependent kinases phosphorylate Rb to control its activity and they are, in turn, specifically inhibited by the Ink4 family of cyclin-dependent kinase inhibitors (CDKIs) which cause arrest at the G1 phase of the cell cycle. Mutations in Rb, cyclin D1, its catalytic subunit Cdk4, and the CDKI p16Ink4a, which alter the protein or its level of expression, are all strongly implicated in cancer. This suggests that the Rb ‘pathway’ is of particular importance1. Here we report the structure of the p19Ink4d protein, determined by NMR spectroscopy2,3,4. The structure indicates that most mutations to the p16Ink4a gene, which result in loss of function, are due to incorrectly folded and/or insoluble protein5. We propose a model for the interaction of Ink4 proteins with D-type cyclin-Cdk4/6 complexes that might provide a basis for the design of therapeutics against cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sherr, C. J. Cancer cell cycles. Science 274, 1672–1677 (1996).
Chan, F. K. M., Zhang, J., Cheng, L., Shapiro, D. N. & Winoto, D. A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16INK4. Mol. Cell. Biol. 15, 2682–2688 (1995).
Guan, K. et al. Isolation and characterization of p19INK4d, a p16 related inhibitor specific to CDK6 and CDK4. Mol. Biol. Cell 7, 57–70 (1996).
Hirai, H., Roussel, M. F., Kato, H.-Y., Ashmun, R. A. & Sherr, C. J. Novel INK4 proteins, p19 and p18, are specific inhibitors of the cyclin D-dependent kinases CDK4 and CDK6. Mol. Cell. Biol. 15, 2672–2681 (1995).
Zhang, B. & Peng, Z.-y. Defective folding of mutant p16INK4a proteins encoded by tumor-derived alleles. J. Biol. Chem. 271, 28734–28737 (1996).
Serrano, M., Hannon, G. J. & Beach, D. Anew regulatory motif in cell-cycle control causing specific inhibition of cyclin D/Cdk4. Nature 366, 704–707 (1993).
Bork, P. Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally. Prot. Struct. Funct. Genet. 17, 363–374 (1993).
Hannon, G. J. & Beach, D. p15INK4B is a potential effector of TGF-β-induced cell-cycle arrest. Nature 371, 257–261 (1994).
Foulkes, W. D., Flanders, T. Y., Pollock, P. M. & Hayward, N. K. The CDKN2A (p16) gene and human cancer. Mol. Med. 3, 5–20 (1997).
Serrano, M. et al. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85, 27–37 (1996).
Merlo, A. et al. 5′ CPG island methylation is associated with transcriptional silencing of the tumor-suppressor p16CDKN2/MTS1 in human cancers. Nature Med. 1, 686–692 (1995).
Yasukawa, T. et al. Increase of solubility of foreign proteins in Escherichia coli by co-production of the bacterial thioredoxin. J. Biol. Chem. 270, 25328–25331 (1995).
Wick, S. T., Dubay, M. M., Imanil, I. & Brizuela, L. Biochemical and mutagenic analysis of the melanoma tumor-suppressor gene-product p16. Oncogene 11, 2013–2019 (1995).
Wölfel, T. et al. Ap16INK4A-insensitive CDK4 mutant targeted by cytolytic T-lymphocytes in a human melanoma. Science 269, 1281–1284 (1995).
Kalus, W. et al. NMR structural characterization of the CDK inhibitor p19INK4d. FEBS Lett. 401, 127–132 (1997).
Gorina, S. & Pavletich, N. P. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science 274, 1001–1005 (1996).
Tevelev, A. et al. Tumor suppressor p16INK4A: Structural characterization of wild-type and mutant proteins by NMR and circular dichroism. Biochemistry 35, 9475–9487 (1996).
Yang, R., Gombart, A. F., Serrano, M. & Koeffler, H. P. Mutational effects on the p16INK4a tumor suppressor protein. Cancer Res. 55, 2503–2506 (1995).
Ranade, K. et al. Mutations associated with familial melanoma impair p16INK4 function. Nature Genet. 10, 114–116 (1995).
Koh, J., Enders, G. H., Dynlacht, B. D. & Harlow, E. Tumour-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature 375, 506–510 (1995).
Parry, D. & Peters, G. Temperature-sensitive mutants of p16CDKN2 associated with familial melanoma. Mol. Cell. Biol. 16, 3844–3852 (1996).
Fåhraeus, R., Paramio, J. M., Ball, K. L., Lain, S. & Lane, D. P. Inhibition of pRB phosphorylation and cell-cycle progression by a 20-residue peptide derived from p16CDKN2/INK4A. Curr. Biol. 6, 84–91 (1996).
Smith, B. O. et al. An approach to global fold determination using limited NMR data from larger proteins selectively protonated at specific residue types. J. Biomol. NMR 8, 360–368 (1996).
Clowes, R. T., Crawford, A., Raine, A. R. C., Smith, B. O. & Laue, E. D. Structural studies of proteins using NMR spectroscopy. Curr. Opin. Biotechnol. 6, 81–88 (1995).
Brünger, A. T. X-PLOR, Version 3.1: A System for X-ray Crystallography and NMR (Yale Univ. Press, New Haven and London, (1992)).
Clore, G. M. & Gronenborn, A. M. Structures of larger proteins in solution—3-dimensional and 4-dimensional heteronuclear NMR spectroscopy. Science 252, 1390–1399 (1991).
Vuister, G. W. & Bax, A. Quantitative J correlation: A new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777 (1993).
Nilges, M., Marcias, M. J., O'Donoghue, S. I. & Oschkinat, H. Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from β-spectrin. J. Mol. Biol. 269, 408–422 (1997).
Kraulis, P. J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Gilson, M. K., Sharp, K. A. & Honig, B. Calculation of the total electrostatic energy of a macromolecular system—solvation energies, binding energies and conformational analysis. J. Comput. Chem. 9, 327–335 (1988).
Acknowledgements
We thank C. J. Sherr for the plasmid expressing GST-p19Ink4d; S. Ishii for plasmids expressing GroEL and GroES; W. Boucher for computer programming; M. Nilges for protocols for the structure calculations; J. Krywko for the model of Cdk4; M. Serrano and D. Beach for assaying p19Ink4d and for communicating unpublished results; N. Pavletich for the coordinates of 53BP2; and A. Murzin for helpful discussion. This work was supported by a grant from the BBSRC. The Cambridge Centre for Molecular Recognition is supported by the BBSRC and the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luh, F., Archer, S., Domaille, P. et al. Structure of the cyclin-dependent kinase inhibitor p19Ink4d. Nature 389, 999–1003 (1997). https://doi.org/10.1038/40202
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/40202
This article is cited by
-
Circular RNA circCSPP1 promotes the occurrence and development of colon cancer by sponging miR-431 and regulating ROCK1 and ZEB1
Journal of Translational Medicine (2022)
-
A dual-reporter system for investigating and optimizing protein translation and folding in E. coli
Nature Communications (2021)
-
Renewal and preliminary study of expressed sequence tags database on human fetal liver aged 22 wk of gestation
Science Bulletin (2008)
-
Germline splicing mutations of CDKN2A predispose to melanoma
Oncogene (2003)
-
Lack of germline CDK6 mutations in familial melanoma
Oncogene (2000)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.