Abstract
A subgroup of testicular seminomas has been reported to contain activating mutations in KIT, the transmembrane tyrosine kinase receptor encoded by the c-kit gene. Most mutations are in exon 17, although exon 11-activating mutations have recently been described. For patients refractory to standard therapeutic protocols for seminoma, the presence of c-kit-activating mutations in some of these neoplasms might suggest an alternative therapy with KIT targeting drugs. We used the novel mutation scanning technique, high-resolution melting amplicon analysis, to screen a series of 22 testicular seminomas for c-kit-activating mutations. Four cases (18%) had exon 17-activating mutations and these included D816Y, D816V, Y823N and one case that contained both D816E and D820H. A single case (5%) had an exon 11-activating mutation. Interestingly, the exon 11-activating mutation was L576P, the same mutation that characterizes the rare c-kit mutation-positive cases of malignant melanoma. Fluorescence in situ hybridization (FISH) for c-kit suggested that most seminomas are probably polysomic for c-kit and there was not a significant difference in c-kit FISH characteristics between the mutation-positive and mutation-negative cases. The use of high-resolution melting amplicon analysis as a screening technique will allow for the rapid identification of patients with testicular seminomas whose tumors contain c-kit-activating mutations. This could benefit patients whose tumors are refractory to standard therapeutic protocols.
Similar content being viewed by others
Main
The product of the c-kit gene, KIT, is a transmembrane receptor tyrosine kinase. Upon ligand binding, KIT undergoes dimerization and activation. Activated KIT then initiates a signaling pathway that leads to antiapoptosis and cell proliferation.1, 2 A functional c-kit gene is necessary for proper development and migration of the interstitial cells of Cajal, blood cell progenitors, mast cells, melanocytes, as well as for normal spermatogenesis.3, 4, 5, 6 It is becoming apparent that many human malignancies that develop from cells with KIT-expressing lineages contain activating mutations in the c-kit gene.7, 8 The c-kit-activating mutations appear restricted to those in exons 9, 11, 13 and 17 and consist of in-frame deletions and insertions as well as point mutations. The success of imatinib (Gleevec) in the treatment of gastrointestinal stromal tumors (GISTs) with c-kit-activating mutations underlies the importance of detecting these alterations in human malignancies.9
Not surprisingly, c-kit-activating mutations have been reported to be present in testicular seminomas.10, 11 Most reports describe point mutations in exon 17, but occasional exon 11 mutations have also been noted.12, 13, 14, 15 The frequency of these mutations in seminomas has been reported to range up to 41%.14 The majority of the exon 17 mutations reported (D816V) code for a c-kit protein that is probably imatinib resistant.16 However, newer agents are being developed that might specifically target this imatinib-resistant c-kit protein17 and if these newer drugs become clinically useful, they might have importance in treating chemotherapeutically refractory seminomas.
In spite of several reports indicating the presence of c-kit mutations in seminomas, a recent comprehensive survey of c-kit mutations in over 3000 tumors from 120 different categories failed to detect any c-kit mutations in seminomas18 and calls into question many of the reports indicating that c-kit mutations characterize a subset of seminomas.
We have recently used the novel mutation screening technology referred to as high-resolution melting amplicon analysis to screen for c-kit-activating mutations in GISTs.19 Because of the potential importance of c-kit-activating mutations in seminomas and because of the discrepancy concerning their frequency, we screened 22 seminomas with high-resolution melting for the presence of c-kit-activating mutations in exons 11 and 17. Positive cases were followed up by direct DNA sequencing. Our results confirm the presence of c-kit-activating mutations in testicular seminomas.
Materials and methods
Sources of Tissue
Paraffin blocks from 22 cases diagnosed as primary testicular seminomas were retrieved from the surgical pathology files at the University of Utah Health Sciences Center. All cases were reviewed to confirm the diagnosis of a seminoma. Only pure seminomas were evaluated and no mixed germ cell tumors were included in the study. Only cases that contained areas enriched in tumor (at least 50% tumor) accessible for microdissection were chosen for study. The use of human tissue for this analysis was approved by the Institutional Review Board (IRB# 11903) at the University of Utah.
Immunohistochemistry
Antibodies against KIT (CD 117) were used at a dilution of 1:400 and were obtained from DAKO Corporation (Carpinteria, CA, USA). Immunohistochemical staining was performed with the Nexes Instrument from Ventana Inc. (Tucson, AZ, USA) in accordance with the manufacturer's instructions and as described previously.19 The chromogen was diaminobenzidine. No antigen retrieval techniques were used. Positive KIT staining mast cells in the testicular parenchyma served as internal controls. Interpretation of the KIT immunostain was performed on a scale of 0 to 3+ as follows: 3+, strong, intense membrane staining; 2+, moderate membrane staining; 1+, weak membrane staining; 0, no detectable to barely discernable membrane staining. All immunostains were interpreted by a single investigator (JAH).
Chemicals, DNA and Enzymes
These materials were described previously.19, 20
Design of Primers
Primers specific for c-kit exons 11 and 17 were designed with the use of Primer Designer Software (Scientific and Education Software, Durham, NC, USA) and have been described previously.19
DNA Isolation from Paraffin-Embedded Tissue
An appropriate paraffin block containing tumor tissue was selected for analysis after reviewing the hematoxylin and eosin (H and E)-stained slides. An area of tumor on the H and E slide was identified on a corresponding unstained slide and circled with a fine tipped pen. Because seminomas generally contain an abundant lymphocytic infiltrate, care was taken to insure that the area chosen for study contained at least 50% tumor as estimated by visual inspection of the slide. Material was scraped from the circled area on the unstained slide and DNA isolated with a proteinase K digestion according to procedures described previously.19
Polymerase Chain Reaction
Polymerase chain reaction (PCR) was performed directly on the proteinase K digest (see above) with the Light Cycler (Roche Diagnostics, Indianapolis, IN, USA) as described previously.19, 20 All reactions contained dUTP in place of dTTP so that incubation of the reactions with uracil N-glycosylase before PCR prevents ‘carry over’ contamination. Genomic DNA was used as a control. All samples were run in triplicate.
High-Resolution Amplicon Melting
After PCR, the samples were momentarily heated to 95°C and then cooled to 40°C. High-resolution amplicon melting analysis of the PCR products was performed with the use of the HR-1 instrument (Idaho Technology, Salt Lake City, UT, USA) as described.19 Mixing experiments with mutant and wild-type DNA indicate that for single base-pair changes, at least 50% of the total DNA isolated should be derived from the mutant in order to easily detect the mutation by melting curve analysis (data not shown).
DNA Sequencing
DNA sequencing and analysis was performed as described previously.19 Several previous studies indicate that a normal melting curve obtained by high-resolution melting amplicon analysis on PCR-amplified products reflects an underlying normal DNA sequence.19, 20, 21 In screening for exon 17 mutations, all cases were followed up by direct bi-directional DNA sequencing. All normal melting curves were found to be the result of normal DNA sequences and all abnormal curves were found to be the result of underlying mutations. Therefore, for exon 11 screening, only the abnormal curves were subjected to follow-up by direct DNA sequencing. The normal melting curves for exon 11 were assumed to reflect an underlying normal DNA sequence and were not studied further.
c-kit Fluorescence In Situ Hybridization
A fluorescence in situ hybridization (FISH) probe to detect the c-kit gene was developed from Escherichia coli containing bacterial artificial chromosome (BAC) c-kit sequences as described.22 The purified c-kit BAC DNA was labeled with the use of the Vysis (Downers Grove, IL, USA) nick translation kit as detailed in the manufacturer's instructions. Nick translation was accomplished in the presence of SpectrumOrange dUTP. To determine the gene copy number of the c-kit gene, the SpectrumOrange-labeled c-kit gene probe was combined with a SpectrumGreen CEP4 probe obtained from Vysis (Downers Grove, IL, USA), and FISH was performed as described recently.22 At least 40 non-overlapping interphase nuclei were counted and the number of orange and green signals recorded. Cases in which the ratio of c-kit signals to CEP4 signals was greater than 2 were interpreted as amplified. Cases in which the CEP4 signals were in the range of 1.76–2.25 signals per cell were interpreted as disomy. Cases that showed CEP4 signals greater than 2.25 were considered polysomy.23
Results
Immunohistochemical Staining for c-kit Expression in Seminomas
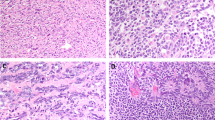
Of the 22 seminomas, 19 (86%) showed at least some membrane staining for KIT. Of the positive staining cases (n=19), six were interpreted as 3+ (32%), eight were interpreted as 2+ (42%) and five (26%) were interpreted as 1+. A seminoma with strong 3+ KIT membrane staining is shown in Figure 1. In many cases, the surrounding seminiferous tubules contained evidence of intratubular germ cell neoplasia, and the malignant cells in these tubules were also strongly KIT positive (Figure 1). Normal seminiferous tubules did not contain cells expressing KIT. This suggests that increased expression of KIT probably most likely occurs in the transformation of testicular germ cells before the development of an invasive tumor.
Expression of KIT in seminoma and intratubular germ cell neoplasia. Immunohistochemical staining for KIT was performed as described in Materials and methods. (a) Testicular seminoma, H and E stain (× 400). (b) KIT stain of testicular seminoma. The KIT stain shows strong membrane staining and was interpreted as 3+ on a scale of 0 (no staining) to 3+ (strong membrane staining) (× 250). (c) Seminiferous tubule with intratubular germ cell neoplasia (× 400). (d) KIT stain of intratubular germ cell neoplasia. The intratubular malignant cells show strong positive (3+) KIT staining (× 400). Normal seminiferous tubules did not show KIT staining (not shown).
High-Resolution Melting Amplicon Analysis for c-kit Exons 11 and 17
All 22 cases of seminoma were screened for c-kit alterations in exons 11 and 17. One case (4.5%) showed an abnormal melting curve for exon 11. Five of the 22 cases (23%) showed an abnormal exon 17 melting curve. Examples of abnormal melting curves are shown in Figure 2.
High-resolution melting amplicon analysis of testicular seminomas. High-resolution melting amplicon analysis was performed as described in Materials and methods. (a) Melting analysis for c-kit exon 11. The DNA melting analysis shows an abnormal melting curve for the tumor (curve B) and DNA sequence analysis indicated a c-kit exon 11 point mutation (L576P). The melting curve obtained from normal genomic DNA is indicated as curve A. (b) Melting analysis for c kit exon 17. The DNA melting analysis shows an abnormal melting curve obtained from DNA isolated from the tumor (curve B). DNA sequence analysis indicated that the alteration was a point mutation in exon 17 (Y823N). The melting curve of normal genomic DNA is indicated as curve A.
Direct DNA Sequencing
Cases showing abnormal melting curves were followed up by direct bi-directional DNA sequencing. For the five cases that showed abnormal melting curves for exon 17, one case contained a D816Y mutation, one case contained a D816V mutation, one case contained both D816E and D820H mutations, one case contained a Y823N mutation in addition to a silent mutation at position 798 (ATC to ATT, I798I) and one case contained just the silent mutation at amino-acid residue position 798. The one case with an abnormal melting curve for c-kit, exon 11 was found to contain an L576P mutation. Therefore, five of the 22 cases (23%) contained activating c-kit mutations.
c-kit FISH
An increased copy number of c-kit has been reported in semimona.24 To determine if the seminomas with c-kit mutations preferentially show increases in c-kit copy number, cases that showed KIT expression (n=19) were analyzed by c-kit FISH. As shown in Table 1, none of the cases with c-kit mutations appeared to amplify the gene. They all appeared polysomic. The average c-kit copy number for the mutation-positive cases was 3.6±2.2 (range 1.80–7.24) and the average chromosome 4 copy number was 3.2±1.6 (range 2.36–5.96). The average c-kit/CEP4 ratio of the mutant cases was therefore 1.1. For the mutation-negative cases, FISH could be performed on 14 of them. The average c-kit copy number of those cases was 3.0±1.8 (range 1.68–8.13) and the average chromosome 4 copy number was 2.43±0.35 (range 1.7–2.88) (data not shown). Thus, many of the non-mutant seminomas were also polysomic. The c-kit/CEP4 ratio in the non-mutation seminomas was 1.2 and only one of the non-mutant seminomas showed c-kit gene amplification with a c-kit/CEP=3.19 (c-kit=8.13, CEP4=2.55) (data not shown). c-kit FISH results were not remarkably different between the mutation-positive and mutation-negative cases.
Discussion
Although a recent report failed to detect c-kit mutations in seminomas,18 other reports have.10, 11, 12, 13, 14, 15 To investigate this discrepancy and to further characterize the types of c-kit mutations in seminomas, we used a new methodology referred to as high-resolution melting amplicon analysis, to screen 22 pure testicular seminomas for c-kit-activating mutations. After PCR amplification of genomic DNA from a tumor with a c-kit-activating mutation, a mixture of DNA molecules is obtained. Some contain the mutation and some do not. Denaturation and re-annealing of the PCR-amplified products leads to formation of heteroduplexes in which one DNA strand contains the mutation and the other does not. Because of the base-pair mismatch(es) in the heteroduplexes, the melting curve of the PCR-amplified DNA will be lower than normal and is easily detectable by melting analysis with high-resolution methodology. This is fundamentally different from other techniques of c-kit mutation scanning such as denaturing high-performance liquid chromatography,11 denaturing gradient gel electrophoresis25 or single-stranded conformational polymorphism.26 In these techniques, mutation detection requires separation and visualization of the heteroduplexes by chromatography or electrophoresis through a solid matrix. Melting analysis does not require separation and visualization of heteroduplexes. Using melting analysis, we were able to detect and subsequently sequence confirm the presence of c-kit mutations in five of 22 seminomas. Therefore, our analysis, with a fundamentally different type of methodology, agrees with many of the reports describing c-kit mutations in seminomas.
Most of our c-kit-activating mutations were in exon 17, which is in agreement with others. Although three of our reported mutations have not been described previously (D816E, D820H and Y823N), these mutations are clustered in the same area as other known c-kit exon 17 mutations and presumably also code for a constitutively activated protein. Analysis of normal tissue from these cases did not reveal the mutation, so presumably these represent somatic mutations in the tumor (C. Willmore-Payne et al, unpublished data). Interestingly, our one c-kit exon 11 mutation was L576P, which has also been observed by others in seminomas.14 c-kit L576P mutations are also present in other tumor systems and are the only c-kit-activating mutations so far reported in melanoma.22 Although the L576P mutation makes up less than 10% of c-kit-activating mutations in GISTs,9 its frequent occurrence as a common c-kit-activating mutation in non-GIST tumors is intriguing.
With standard therapeutic protocols, the cure rate of testicular seminomas approaches 90%.27 Most of the c-kit-activating mutations in seminomas are in exon 17, which is generally regarded as coding for an imatinib-resistant protein. However, new drugs directly targeting imatinib-resistant KIT proteins are being developed. These new drugs might find use in treating refractory seminomas. The rare seminoma with an exon 11 mutation would be predicted to be imatinib sensitive. If the use of c-kit-targeting drugs becomes a reality in the therapy of testicular seminomas, high-resolution melting amplicon analysis could be used to help identify a potential group of responsive patients.
References
Roskoski Jr R . Structure and regulation of Kit protein-tyrosine kinase—the stem cell factor receptor. Biochem Biophys Res Commun 2005;338:1307–1315.
Roskoski Jr R . Signaling by Kit protein-tyrosine kinase—the stem cell factor receptor. Biochem Biophys Res Commun 2005;337:1–13.
Tan JC, Nocka K, Ray P, et al. The dominant W42 spotting phenotype results from a missense mutation in the c-kit receptor kinase. Science 1990;247:209–212.
Gibson PC, Cooper K . CD117 (KIT): a diverse protein with selective applications in surgical pathology. Adv Anat Pathol 2002;9:65–69.
Hoyer PE, Byskov AG, Mollgard K . Stem cell factor and c-Kit in human primordial germ cells and fetal ovaries. Mol Cell Endocrinol 2005;234:1–10.
Bedell MA, Mahakali Zama A . Genetic analysis of Kit ligand functions during mouse spermatogenesis. J Androl 2004;25:188–199.
Heinrich MC, Blanke CD, Druker BJ, et al. Inhibition of KIT tyrosine kinase activity: a novel molecular approach to the treatment of KIT-positive malignancies. J Clin Oncol 2002;20:1692–1703.
Miettinen M, Lasota J . KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol 2005;13:205–220.
Corless CL, Fletcher JA, Heinrich MC . Biology of gastrointestinal stromal tumors. J Clin Oncol 2004;22:3813–3825.
Tian Q, Frierson Jr HF, Krystal GW, et al. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol 1999;154:1643–1647.
Kemmer K, Corless CL, Fletcher JA, et al. KIT mutations are common in testicular seminomas. Am J Pathol 2004;164:305–313.
Sakuma Y, Sakurai S, Oguni S, et al. Alterations of the c-kit gene in testicular germ cell tumors. Cancer Sci 2003;94:486–491.
Sakuma Y, Sakurai S, Oguni S, et al. c-kit gene mutations in intracranial germinomas. Cancer Sci 2004;95:716–720.
Nakai Y, Nonomura N, Oka D, et al. KIT (c-kit oncogene product) pathway is constitutively activated in human testicular germ cell tumors. Biochem Biophys Res Commun 2005;337:289–296.
Pauls K, Wardelmann E, Franke FE, et al. Primary extragonadal germ cell tumour: unusual localization of a c-kit mutated retroperitoneal seminoma in the gastric wall. Histopathology 2005;47:111–119.
Ma Y, Zeng S, Metcalfe DD, et al. The c-KIT mutation causing human mastocytosis is resistant to STI571 and other KIT kinase inhibitors: kinases with enzymatic site mutations show different inhibitor sensitivity profiles than wild-type kinases and those with regulatory-type mutations. Blood 2002;99:1741–1744.
Schittenhelm MM, Shiraga S, Schroeder A, et al. Dasatinib (BMS-354825), a dual SRC/ABL kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant KIT isoforms associated with human malignancies. Cancer Res 2006;66:473–481.
Went PT, Dirnhofer S, Bundi M, et al. Prevalence of KIT expression in human tumors. J Clin Oncol 2004;22:4514–4522.
Willmore C, Holden JA, Zhou L, et al. Detection of c-kit-activating mutations in gastrointestinal stromal tumors by high-resolution amplicon melting analysis. Am J Clin Pathol 2004;122:206–216.
Willmore-Payne C, Holden JA, Tripp S, et al. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol 2005;36:486–493.
Willmore-Payne C, Holden JA, Layfield LJ . Detection of EGFR- and HER2-activating mutations in squamous cell carcinoma involving the head and neck. Mod Pathol 2006;19:634–640.
Willmore-Payne C, Holden JA, Hirschowitz S, et al. BRAF and c-kit gene copy number in mutation positive malignant melanoma. Hum Pathol 2006;37:520–527.
Mezzelani A, Alasio L, Bartoli C, et al. c-erbB2/neu gene and chromosome 17 analysis in breast cancer by FISH on archival cytological fine-needle aspirates. Br J Cancer 1999;80:519–525.
McIntyre A, Summersgill B, Grygalewicz B, et al. Amplification and overexpression of the KIT gene is associated with progression in the seminoma subtype of testicular germ cell tumors of adolescents and adults. Cancer Res 2005;65:8085–8089.
Beghini A, Tibiletti MG, Roversi G, et al. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer 2001;92:657–662.
Koay MH, Goh YW, Iacopetta B, et al. Gastrointestinal stromal tumours (GISTs): a clinicopathological and molecular study of 66 cases. Pathology 2005;37:22–31.
Carver BS, Sheinfeld J . Germ cell tumors of the testis. Ann Surg Oncol 2005;12:871–880.
Acknowledgements
We thank Trena Held for preparing the table. We also thank Dr Carl Wittwer for his support and help with high-resolution melting analysis. This work was supported by the Associated Regional and University Pathologists (ARUP) Institute for Clinical and Experimental Pathology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Willmore-Payne, C., Holden, J., Chadwick, B. et al. Detection of c-kit exons 11- and 17-activating mutations in testicular seminomas by high-resolution melting amplicon analysis. Mod Pathol 19, 1164–1169 (2006). https://doi.org/10.1038/modpathol.3800623
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800623
Keywords
This article is cited by
-
The stem cell factor (SCF)/c-KIT signalling in testis and prostate cancer
Journal of Cell Communication and Signaling (2017)
-
High-resolution melting analysis for rapid screening of BRCA1 and BRCA2 Spanish mutations
Breast Cancer Research and Treatment (2009)
-
Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis
Nature Protocols (2007)
-
A subset of colorectal carcinomas express c-KIT protein independently of BRAF and/or KRAS activation
Virchows Archiv (2007)
-
High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer
BMC Cancer (2006)