Abstract
Carcinosarcomas and carcinoma ex pleomorphic adenoma of the salivary glands are rare tumors that fit into the broader category of malignant mixed tumors. Although most evidence has suggested that the different morphologic components arise from a common clonal origin, there are very few studies that have provided molecular evidence for this clonality. In this study, we examined a set of seven carcinosarcomas and four carcinomas ex pleomorphic adenoma for tumor suppressor gene loss of heterozygosity, in order to assess the clonal patterns in the varying components. Microdissection was performed to obtain each morphological component and tumor suppressor gene loci on 3p, 5q, 9p, 17p, 17q, and 18q were analyzed. The fractional allelic loss (FAL) was calculated for each area, and the different targets were compared for their molecular profile. The overall mean FAL of the malignant targets was 42%. In carcinosarcomas, the sarcomatous targets had a higher mean FAL than the carcinomatous targets (68 vs 46%, respectively) and in carcinomas ex pleomorphic adenoma, the mean FAL in the benign component was 11 vs 46% seen in the carcinomatous component. The most frequently lost genetic loci were p53 (17p13, 73%), nm23-H1 (17q21, 55%), and DCC (18q21, 50%). Loss of heterozygosity of 17q21 and 9p21 only occurred in carcinosarcomas and not in carcinomas ex pleomorphic adenoma. Within the carcinosarcomas, the mutational profiles were conserved between epithelial and sarcomatous areas. In carcinomas ex pleomorphic adenoma, loss of heterozygosity was uncommon in the benign component, but the mutations were conserved in the corresponding malignant areas. These results support the hypothesis that the carcinomatous and sarcomatous components of carcinosarcomas are clonally related. Furthermore, these data support prior studies that suggest a common clonal origin for the benign and malignant components of carcinomas ex pleomorphic adenoma.
Similar content being viewed by others
Main
Malignant mixed tumors of the salivary gland are diverse and include three different subcategories of lesions: carcinosarcomas, carcinoma ex pleomorphic adenoma, and metastasizing pleomorphic adenomas. Both types of frankly malignant tumors behave aggressively, with poor 5-year survival.1, 2
Carcinosarcomas (true malignant mixed tumors) of the salivary glands are rare biphasic tumors that exhibit both carcinomatous and sarcomatous elements.3 These tumors are thought to develop de novo in the salivary gland, and contain malignant stromal and epithelial elements.4 The carcinomatous component varies, but can be composed of adenocarcinoma, squamous cell carcinoma, or undifferentiated carcinoma. They may also show specific salivary carcinoma phenotypes, including salivary duct carcinoma or adenoid cystic carcinoma. Sarcomatous elements can also be variable, including chondrosarcoma, osteosarcoma, fibrosarcoma, and malignant fibrous histiocytoma. Most frequently, the sarcomatous component dominates, though the two elements can be found in an intermixed pattern.5
Carcinoma ex pleomorphic adenoma arises from a pre-existing benign mixed tumor. Usually, the patient will have a long-standing history of a salivary gland lesion, and may have undergone multiple operations for recurrent pleomorphic adenoma. Most of these tumors will have malignant epithelial component, but not malignant stromal components. Based on the morphologic features and few genetic analyses, most investigators have hypothesized that the carcinoma arises directly from de-differentiation or malignant transformation of the benign tumor.6, 7
In carcinosarcomas, however, the relationship between the different elements is controversial.8 Do the two morphologically distinct elements arise from a common precursor cell or do they arise separately and represent a collision tumor? In other organ systems, the two components of biphasic carcinosarcomas have been genetically linked to one another.9, 10 Very few molecular studies have addressed this question in the salivary gland, but those that have suggest that the sarcomatous and carcinomatous elements are pathogenetically linked.11, 12
In this study, we studied a group of malignant mixed tumors, including both carcinosarcomas and carcinoma ex pleomorphic adenomas of salivary gland with a loss of heterozygosity analysis of a panel of tumor suppressor genes. We describe the mutational profile for each morphologically unique tumor component, with direct comparison between the different compartments, as a tool to assess common clonal origin.
Materials and methods
Inclusion criteria for this study were the diagnoses of malignant mixed tumor of salivary gland origin. Only cases with blocks and slides available were included. Tumors were selected from the files of the University of Pittsburgh and West Virginia University Divisions of Surgical Pathology. The diagnosis was confirmed from review of the H&E slides and special stains. Pathologic information was obtained from review of the cases and from the surgical pathology report. Clinical information and follow-up were obtained from the electronic medical record; this was obtained by an honest broker and de-identified. This study was approved by the Institutional Review Board of the University of Pittsburgh and West Virginia University.
Unstained, deparaffinized sections were obtained, with an H&E-stained slide before and after to ensure that the microdissection targets were present on all levels. Microdissection targets were marked on the first H&E-stained slide. The target areas were then microdissected from the unstained slides and DNA was extracted from the resultant tissue fragments after digestion with proteinase K using the Qiagen DNeasy extraction kit (Qiagen, Valencia, CA, USA).
PCR was performed using a set of primers that amplify short tandem repeat units that are near-known tumor suppressor genes (Table 1). The PCR primers are designed to give relatively small product sizes (optimally <200 basepairs), as tissue that has been fixed in formalin has unavoidable DNA degradation. The PCR products were then analyzed using capillary electrophoresis and Genescan software for fragment analysis (ABI Prism 3100, Applied Biosystems Inc., Foster City, CA, USA). The ratio of the peak heights (allele ratio) was obtained for informative (heterozygous) markers in both the tumor and the normal tissues. The allele ratio for tumor was divided by the allele ratio for normal in order to assess for loss of heterozygosity. Ratios over 2.0 and under 0.5 were considered to be indicative of LOH in the target being analyzed.13 The fractional allelic loss (FAL) was calculated (number of mutations detected divided by the total number of informative loci).14 Statistical analysis was performed using SPSS software (χ2 and Student's T-test).
Results
A total of 13 different specimens of malignant mixed tumors arising in 11 different patients were included in this study. The tumors included six high-grade carcinosarcoma, three recurrences of a low-grade carcinosarcoma (same patient), and four carcinoma ex pleomorphic adenoma. No metastasizing pleomorphic adenomas had been diagnosed at either institution participating in this study and sarcomatoid salivary duct carcinomas were not included. The patients had an average age of 67 years (range 40–90). There were four females and seven males. Most of the tumors arose in the parotid gland (81%). The clinical characteristics of the patients in this series are shown in Table 2.
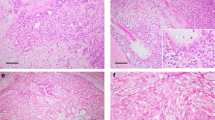
Among the carcinosarcomas, the malignant stromal component was composed of undifferentiated spindle cells and the malignant epithelial component was poorly differentiated adenocarcinoma in all cases (Figures 1 and 2). These cases did not have definitive features or the staining pattern of salivary duct carcinoma. The low-grade carcinosarcoma showed a stromal component that was hypercellular, but relatively bland and without abundant mitoses. The carcinoma ex pleomorphic adenomas showed adenocarcinoma and there was a recognizable benign pleomorphic adenoma as a separate component (Figures 3 and 4).
The malignant tumor components analyzed at the molecular level all had a relatively high frequency of allelic loss, with an average FAL of 42% for all malignant targets examined. Interestingly, allelic loss of 17q21 and 9p21 were only seen in carcinosarcomas and not in carcinoma ex pleomorphic adenomas. Allelic losses in three genetic loci occurred at high rates: 17p13, which is the location of the p53 gene (73% of targets), 17q, in the location of the nm-23 gene (55% of targets), and 18q, in the location of the DCC gene (50% of targets).
In comparing the genotype between morphologically distinct areas within one tumor, we found that there was excellent concordance between the malignant carcinomatous and sarcomatous elements in carcinosarcomas. Overall, 73% of the loci tested in these components showed concordant mutational profiles. The stromal component showed higher FALs in most cases (68% FAL in stromal elements vs 46% FAL in carcinomatous elements), but this did not reach statistical significance. In the carcinoma ex pleomorphic adenomas, there were few allelic losses seen in the benign component (mean 11%), and a significantly higher rate of allelic loss in the malignant areas (mean 46%), (P=0.01). Allelic losses were seen in the benign pleomorphic component in only two of four cases, and these were conserved in the malignant component.
The malignant epithelial components in carcinosarcomas had significantly higher FAL than the malignant epithelial component from carcinoma ex pleomorphic adenoma (55 vs 17%, P=0.001). Additionally, the FAL was lower in the one low-grade carcinosarcoma as compared to the mean FAL of the histologically high-grade tumor areas in the carcinosarcomas (mean FAL for all targets in low-grade tumor 7.2% vs mean FAL for all targets in high-grade tumors 45%).
Discussion
Malignant mixed tumors of salivary gland origin are rare, accounting for <1% of overall tumors of the salivary glands.1 They can be divided into three general categories, the first two of which show frankly malignant histologic features: carcinosarcoma, carcinoma ex pleomorphic adenoma, and metastasizing pleomorphic adenoma. Although the existence of the latter category has been disputed, it is recognized in the WHO classification, and is described as having a 40–50% mortality rate.15
Carcinosarcomas are tumors that contain both malignant stromal components and malignant epithelial components. These elements can have a specific morphologic differentiation, with recognizable histologic features, or they can be undifferentiated malignant tumors that only retain the basic stromal or epithelial nature. These tumors are thought to most commonly arise de novo and not from a pre-existing benign tumor.
Carcinoma ex pleomorphic adenomas arise from a pre-existing benign mixed tumor. Many of these patients will have a history of a stable salivary gland tumor or a may have a history of multiple surgeries for recurrent pleomorphic adenoma. Malignant transformation of these tumors is thought to take up to 20 years or more. Both of these tumor types have an aggressive clinical course.8, 16 Both low- and high-grade carcinomas can be present in carcinoma ex pleomorphic adenoma.17
The molecular profiles of malignant mixed tumors of salivary gland origin, and in particular carcinosarcomas, has not been well established. Although it is commonly suggested that the morphologically unique areas in these mixed tumors are related pathogenetically, there have been relatively few studies that attempted to identify the common clonal genetic changes.7, 12
In this study, we examined the mutational profile of a series of malignant mixed tumors using a panel of tumor suppressor genes that have been implicated in the pathogenesis of salivary gland tumors. These tumor suppressor genes included loci on 3p26, 5q23, 9p21, 17p13, 17q21, and 18q21. These loci correspond to the areas that contain the tumor suppressor genes VHL, hOGG1, APC, CDKN2, p53, nm-23, and DCC. By microdissecting the different morphologically distinct components for molecular analysis, we sought to demonstrate clonal genetic changes that were conserved between the different elements. In carcinosarcomas, we sought to demonstrate that malignant epithelial and sarcomatous elements were clonally related; in carcinoma ex pleomorphic adenoma, we sought to demonstrate clonal losses that were conserved between the benign and the malignant components.
We did find that the malignant epithelial and sarcomatous elements in carcinosarcoma shared similar patterns of allelic losses. There were more mutations present in the sarcomatous component, suggesting accumulations of additional genetic hits in these areas. This supports the theories that the different components arise from a common clonal origin and do not represent collision tumors arising from two separate independent malignant processes. The FAL of the low-grade carcinosarcoma studied was lower than that of the higher grade tumors. Interestingly, the low-grade carcinosarcoma had different molecular profiles at the different times it was sampled, suggesting that there is probably some intratumoral heterogeneity.
In the carcinoma ex pleomorphic adenomas that we included in this series, common clonal origin was only suggested for 2/4 cases, in which shared mutations could be shown. The remaining two cases did not have any mutations identified in the pleomorphic adenoma component and therefore a common clonal origin could not be demonstrated. Overall, the FAL of the benign elements was relatively low (mean FAL 11.3%). The malignant components had significantly more allelic losses (mean FAL of malignant component 46%). Our results in at least two cases do support prior work that has identified common clonal origin.7 These low mutational rates also argue against the idea of possibly detecting at risk precursor lesions in otherwise benign pleomorphic adenomas.
In summary, we have demonstrated that the FAL and mutational profiles for the carcinomatous and the sarcomatous areas in salivary gland carcinosarcomas are similar. These data provide supporting evidence for the theory that all malignant elements in these tumors are derived from a common clonal precursor. We have also shown that the benign components of carcinoma ex pleomorphic adenoma do not show high-level frequency of allelic loss. Finally, our results show that the carcinosarcomas have higher overall mutational rates than the carcinoma ex pleomorphic adenomas.
Additional work to demonstrate clonality in the benign and malignant components of carcinoma ex pleomorphic adenomas, such as increasing the number of loci tested to include mutational markers for very early events in neoplasia for pleomorphic adenomas. However, in these cases, the history and the histologic features are often very supportive of a common origin. Finally, additional work with the molecular profiles of malignant mixed tumors to identify any potential prognostic marker would be valuable to optimize patient treatment and management for these aggressive tumors.
References
Wahlberg P, Anderson H, Biorklund A, et al. Carcinoma of the parotid and submandibular glands—a study of survival in 2465 patients. Oral Oncol 2002;38:706–713.
Tortoledo ME, Luna MA, Batsakis JG . Carcinomas ex pleomorphic adenoma and malignant mixed tumors. Histomorphologic indexes. Arch Otolaryngol Head Neck Surg 1984;110:172–176.
Seifert G . Histopathology of malignant salivary gland tumours. Oral Oncol 1992;28B:49–56.
Sironi M, Isimbaldi G, Claren R, et al. Carcinosarcoma of the parotid gland: cytological, clinicopathological and immunohistochemical study of a case. Pathol Res Practice 2000;196:511–517.
Kwon MY, Gu M . True malignant mixed tumor (carcinosarcoma) of parotid gland with unusual mesenchymal component: a case report and review of the literature. Arch Pathol Lab Med 2001;125:812–815.
Lewis JE, Olsen KD, Sebo TJ . Carcinoma ex pleomorphic adenoma: pathologic analysis of 73 cases. Hum Pathol 2001;32:596–604.
El-Naggar AK, Callender D, Coombes MM, et al. Molecular genetic alterations in carcinoma ex-pleomorphic adenoma: a putative progression model? Genes Chromosomes Cancer 2000;27:162–168.
Stephen J, Batsakis JG, Luna MA, et al. True malignant mixed tumors (carcinosarcoma) of salivary glands. Oral Surg Oral Med Oral Pathol 1986;61:597–602.
Matsumoto T, Fujii H, Arakawa A, et al. Loss of heterozygosity analysis shows monoclonal evolution with frequent genetic progression and divergence in esophageal carcinosarcoma. Hum Pathol 2004;35:322–327.
Rossi G, Cavazza A, Sturm N, et al. Pulmonary carcinomas with pleomorphic, sarcomatoid, or sarcomatous elements: a clinicopathologic and immunohistochemical study of 75 cases. Am J Surg Pathol 2003;27:311–324.
Gotte K, Riedel F, Coy JF, et al. Salivary gland carcinosarcoma: immunohistochemical, molecular genetic and electron microscopic findings. Oral Oncol 2000;36:360–364.
Thompson L, Chang B, Barsky SH . Monoclonal origins of malignant mixed tumors (carcinosarcomas). Evidence for a divergent histogenesis. Am J Surg Pathol 1996;20:277–285.
Slebos RJ, Umbach DM, Sommer CA, et al. Analytical and statistical methods to evaluate microsatellite allelic imbalance in small amounts of DNA. Lab Invest 2004;84:649–657.
Nunn J, Scholes AG, Liloglou T, et al. Fractional allele loss indicates distinct genetic populations in the development of squamous cell carcinoma of the head and neck (SCCHN). Carcinogenesis 1999;20:2219–2228.
Barnes L, Eveson J, Reichart PA, et al. Lymphoepithelial carcinoma. In: Kleihues P, Sobin LH (eds). World Health Organization Classification of Tumors: Pathology & Genetics of Head and Neck Tumors. IARC Press: Lyon, 2005, p18.
Felix A, Rosa JC, Fonseca I, et al. Pleomorphic adenoma and carcinoma ex pleomorphic adenoma: immunohistochemical demonstration of the association between tenascin expression and malignancy. Histopathology 2004;45:187–192.
Olsen KD, Lewis JE . Carcinoma ex pleomorphic adenoma: a clinicopathologic review. Head Neck 2001;23:705–712.
Acknowledgements
We thank the members of the Molecular Anatomic Pathology Laboratory for their assistance in performing this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was presented at the United States and Canadian Association for Pathology meeting, Chicago, IL, USA.
Rights and permissions
About this article
Cite this article
Fowler, M., Fowler, J., Ducatman, B. et al. Malignant mixed tumors of the salivary gland: a study of loss of heterozygosity in tumor suppressor genes. Mod Pathol 19, 350–355 (2006). https://doi.org/10.1038/modpathol.3800533
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800533
Keywords
This article is cited by
-
Carcinosarcoma of Submandibular Salivary Gland with a Rare Sarcomatous Variant
Indian Journal of Surgical Oncology (2019)
-
Carcinosarcoma ex Non-Recurrent Pleomorphic Adenoma Composed of TTF-1 Positive Large Cell Neuroendocrine Carcinoma and Myofibrosarcoma: Apropos a Rare Case
Head and Neck Pathology (2013)
-
Carcinoma ex Pleomorphic Adenoma: A Comprehensive Review of Clinical, Pathological and Molecular Data
Head and Neck Pathology (2012)