Abstract
Among the genetic abnormalities reported to occur in mucosa-associated lymphoid tissue (MALT) lymphomas, the three translocations t(11;18)(q21;q21), t(1;14)(p22;q32) and t(14;18)(q32;q21) are of particular interest because they appear to be specific for, or at least closely related to this type of B-cell non-Hodgkin's lymphoma. These translocations affect the MALT1 (18q21) and BCL10 (1p22) genes. We retrieved 77 consecutive biopsies of MALT lymphomas (documented with frozen material) over a 10-year period and investigated these cases for the presence of these three translocations with fluorescence in situ hybridisation, along with the immunohistochemical analysis of the intracellular localisation of the BCL10 protein. The above-listed translocations occurred mutually exclusive and were detected in 10, 1 and 3% of the cases, respectively (the latter incidence being much lower than in the previously reported studies by one single group). These genetic rearrangements corresponded well with the aberrant subcellular localisation of the BCL10 protein as found by immunohistochemistry: t(11;18)(q21;q21) and (1;14)(p22;q32) were marked by a, respectively, moderate to strong nuclear BCL10 staining pattern while t(14;18)(q32;q21)-positive MALT lymphomas were characterised by a perinuclear BCL10 staining pattern. This study further supports the close interaction between the MALT1 and BCL10 proteins in the pathogenesis of MALT lymphomas and may indicate that BCL10 immunohistochemistry is a simple technique to identify those MALT lymphoma cases with an underlying genetic aberration.
Similar content being viewed by others
Main
Extranodal marginal zone lymphoma of the mucosa-associated lymphoid tissue (MALT) or MALT lymphoma is listed as a distinct disease entity in the recently published World Health Organisation (WHO) classification of lymphoid tumour.1 It is characterised by several recurrent chromosomal aberrations including t(1;14)(p22;q32), t(11;18)(q21;q21), t(14;18)(q32;q21), t(3;14)(p13;q32), and by trisomy 3 and 18.
The t(1;14)(p22;q32) results in deregulation of the BCL10 gene after its juxtaposition with the regulatory sequences of the immunoglobulin heavy chain gene cluster (IGH).2 BCL10 encodes a 32 kDa apoptosis regulatory molecule, characterised by an amino-terminal caspase recruit domain (CARD) motif and a Ser/Thr-rich carboxyl terminus of unknown function.3 Monoclonal antibodies raised against the BCL10 protein demonstrate cytoplasmic expression of BCL10 in lymphoid tissue and in breast epithelium.4 In normal lymphoid tissue, BCL10 is expressed exclusively in the cytoplasm of germinal centre and marginal zone B cells. In MALT lymphomas, the BCL10 expression might be observed in the nucleus as well. Nuclear BCL10 expression has been linked to the presence of t(1;14)(p22;q32) or t(11;18)(q21;q21), but the mechanisms of the aberrant nuclear localisation of BCL10 in malignant B cells remain unclear.5, 6
The MALT1 gene is directly involved in the MALT lymphoma-associated chromosomal translocations t(11;18)(q21;q21) and t(14;18)(q32;q21). The first translocation is the most common structural chromosomal abnormality in gastric MALT lymphoma.7, 8 It leads to the generation of a fusion protein comprising the three BIR (baculovirus inhibitor of apoptosis protein repeat) domains present in the N-terminus of the API2 protein and a variable part of the MALT1 protein, which always contains the caspase-like p20 domain.9 The chimeric protein API2-MALT1 effectively activates the NF-κB survival pathway in vitro, but the precise mechanism used to accomplish this activation, as well as the subsequent downstream events that lead to lymphomagenesis, is unknown. The presence of the API2-MALT1 fusion transcript in gastric MALT lymphomas has been linked to resistance to Helicobacter pylori eradication and the absence of transformation to a more aggressive large B-cell lymphoma.10 Moreover, the t(11;18)(q21;q21) appears as a sole chromosomal aberration.11 The t(14;18)(q32;q21) was only very recently identified. Similar to t(1;14)(q21;q21), this translocation is mediated by IGH and leads to overexpression of the MALT1 gene.12, 13 In contrast to t(11;18)(q21;q21)-positive cases, MALT lymphomas marked by t(14;18)(q32;q21) are mainly found outside the gastrointestinal or pulmonary tract, presenting as tumours of the ocular adnexa, skin or salivatory glands. Hypothesis is that this polarisation reflects a different pathogenesis; that is that MALT lymphomas of the salivary and lachrymal glands are often associated with autoimmune disease, whereas those arising in the stomach and lung are linked to an (un)known infectious agent.13
In the present study, we collected 77 consecutive cases of MALT lymphomas. These cases were investigated for the presence of rearrangements of BCL10 and MALT1 by fluorescence in situ hybridisation (FISH) and for the immunohistochemical expression of the BCL10 protein.
Materials and methods
Case Selection
A total of 91 consecutive cases of extranodal marginal zone lymphomas was collected over a 10-year period in the Department of Pathology (University Hospital of K.U. Leuven, Leuven, Belgium). In 77 cases, both fresh frozen tumour tissue and formalin- or B5-fixed paraffin-embedded blocks were available; only these cases were included in the study. Paraffin-embedded material was used for immunohistochemistry and frozen material for FISH analysis. The frozen samples had been stored at −80°C. Among the 77 MALT lymphoma cases, 24 were located in the stomach (14 gastrectomy specimens and 10 endoscopic biopsies), one in the colon (hemicolectomy specimen), seven in the lung (all were lobectomy specimens), three in the nasopharynx, one in the tonsil, 15 in the salivary gland, two in the thyroid gland, 18 in the ocular adnexa, five in the skin and one in the breast. Part of these cases was included in previous studies.5, 7, 9, 14, 15, 16, 17 Gastric endoscopic biopsies were included in this study on condition that clonal Ig heavy chain rearrangement was demonstrated to support the diagnosis of a MALT lymphoma. All cases were reviewed and diagnosed as extranodal marginal zone lymphomas according to the recent criteria of the WHO.1
Immunohistochemistry
Paraffine sections were immunostained for BCL10 (mouse monoclonal anti-human antibody (DakoCytomation, Glosturp, Denmark)). In brief, 4 μm paraffin sections were placed on silanised slides (DakoCytomation), dewaxed in xylene and rehydrated in decreasing concentrations of ethanol. While immersed in citrate buffer (pH 6.0), slides were placed in a calibrated warm water bath (95–99°C) for 3 min to perform epitope retrieval. Incubation with the BCL10 antibody (1:40) was carried out overnight at 4°C. Staining was performed using the EnVision system (DakoCytomation) according to the manufacturer's recommendations. All incubation steps were followed by a wash in three changes of phosphate-buffered saline (pH 7.6). Two splenic biopsies and three lymph node biopsies without obvious abnormalities were used as control tissues.
Cytogenetics and FISH Analysis
Cytogenetic analysis was performed after an overnight culture of tumour specimen according to standard methods. One to 20 G-banded metaphases were analysed. Chromosomal aberrations are presented in accordance with the International System for Human Cytogenetic Nomenclature.18
Interphase FISH analysis was performed on cytospin preparations from single cells isolated from frozen tumour tissue sections (n=55) or from available cytogenetic cells suspensions stored at −20° (n=22). FISH followed the previously published protocols.19, 20 The status of the MALT1 gene was analysed using the LSI MALT1 Dual Color, Break Apart Rearrangement Probe (Vysis, Downers Grove, IL, USA) that consists a mixture of two DNA probes flanking the gene (the first probe, approximately ∼1040 kb in length and labelled in SpectrumGreen™, lies upstream of the MALT1 gene and remains on chromosome 18 in case of a translocation of MALT1; the second, a ∼650 kb probe labelled in SpectrumOrange™, contains sequences derived immediately downstream of the MALT1 gene and is translocated to the partner chromosome in case of a translocation involving MALT1). In normal cells, two fused or colocalised hybridisation signals of LSI MALT1 are seen while in case of MALT1-associated translocation one fused, one red and one green signal are observed. Cases with the latter FISH pattern were further investigated using IGH probes (SO-labelled RP11-11771 and SG-labelled RP11-312H5, kindly provided by T Poulsen, University of Copenhagen, Denmark)21 and API2 (SO-labelled RP11-400E19/RP11-640G3 and SG-labelled RP11-315O6/RP11-10O13) (www.ensembl.org). The status of the BCL10 gene was analysed with a set of BAC clones flanking the gene (SO-labelled RP11-1080I/1RP11-40K4 and SG-labelled RP11-1077C10/RP11-36L4). In case with the BCL10 rearrangement pattern FISH with the IGH probes was applied to confirm the t(1;14)(p22;q32). Evaluation of FISH experiments was performed using a Zeiss Axioplan 2 fluorescence microscope (Zeiss, Zaventem, Belgium). In each experiment 200 cells were examined. The determined cutoff value for the detection of a rearrangement of MALT1, BCL10, IGH and API2 was 6% which is above the mean percentage of cells with a false-positive signal constellation plus 3SDs, as assessed on five lymphoid tissue samples without obvious abnormalities (three lymph nodes biopsies and two splenic biopsies).
Results
Immunohistochemistry
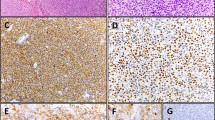
Analysis of the normal spleen and lymph node biopsies demonstrated a weak expression of the BCL10 protein, restricted to the cytoplasm of germinal centre cells and marginal zone cells; in contrast, no BCL10 expression was found in the lymphocytic corona. In none of the normal samples analysed, nuclear staining was observed. A positive BCL10 staining was found in 51 of the 77 MALT lymphomas (66%) while the remaining 26 cases (34%) were completely negative. Among the BCL10-positive lymphoma cases, five patterns of BCL10 expression could be recognised based on its subcellular localisation (Figure 1): an exclusive nuclear BCL10 expression (in nine of the 77 cases or 12%), a combined nuclear and cytoplasmic BCL10 positivity (in 22 of the 77 cases or 29%), a perinuclear BCL10 expression (in two of the 77 cases or 3%), a strong diffuse cytoplasmic BCL10 expression (in five of the 77 cases or 7%), and a weak diffuse cytoplasmic BCL10 expression (in 13 of the 77 cases or 17%). The latter staining pattern did not differ in any way from the BCL10 expression in germinal centre cells and marginal zone cells in control tissues and was therefore not considered to be abnormal.
Cytogenetics
Cytogenetic analysis was performed in 35 cases. In all, 24 cases showed chromosomal aberrations and these cases are listed in Table 1; five cases showed a normal karyotype and in the six remaining cases, analysis was unsuccessful (no mitosis). The most frequent abnormality, gain of chromosome 3 (whole or partial), was detected in 10 cases. Nine cases showed trisomy 18 (one with a sole +18, and four with a simultaneous +3). Translocations t(1;14)(p22;q32), t(11;18)(q32;q21) and t(14;18)(q32;q21) were observed in one, two and one cases, respectively. In four cases other translocations mediated by 14q32/IGH were identified. These translocations include t(3;14)(q27;q32) involving BCL6, t(1;14)(q21;q32), t(3;14)(p13;q32) and t(3;14)(p21;q32). Interestingly, in case no 19 two trisomy 18-positive cell clones, one with t(14;18)(q32;q21) and the second with trisomy 12 and 19, were detected.
FISH
In all, 10 of the 77 cases (13%) contained a significant percentage of nuclei with a signal constellation that indicates a breakpoint in the MALT1 gene locus (Table 2). In eight of these cases, the t(11;18)(q21;q21) was confirmed by the dual-colour break apart probe for API2. From these t(11;18)(q21;q21)-positive cases, six were located in the gastrointestinal tract, while the two other cases arose in the lung and the parotid gland. In the remaining two cases with MALT1 rearrangement (both located in the lachrymal glands), MALT1 was affected by the t(14;18)(q32;q21) as shown by FISH with IGH probes. In 18 cases (23%) an increased number of fused MALT1 signals was found, possibly due to trisomy 18 (16 cases) or polysomy 18 (two cases). Interestingly, one gastric MALT lymphoma case displayed an amplification of the MALT1 gene present in eight to 10 copies per cell. The BCL10 rearrangement mediated by IGH was detected in only one case of a nasopharyngeal lymphoma. In total, 48 MALT lymphomas (62%) did not harbour any FISH detectable aberrations, neither of the BCL10 nor of the MALT1 genes. With the exception of tri- or polysomy 18, the BCL10- and MALT1-related genetic aberrations corresponded well with the BCL10 staining pattern: t(11;18)(q21;q21) and (1;14)(p22;q32) were marked by a, respectively, moderate to strong nuclear BCL10 staining pattern while t(14;18)(q32;q21)-positive MALT lymphomas were characterised by a perinuclear BCL10 staining pattern. However, nuclear (with or without cytoplasmic) BCL10 staining was not specific as it also occurred in eight cases with tri-or polysomy 18, and in 13 cases without detectable BCL10 and MALT1 abnormalities (Table 3).
Discussion
We found an overall incidence of t(11;18)(q32;q21) of 10%, which is in line with other studies that analysed large series of MALT lymphomas for the presence of this translocation by means of FISH, RT-PCR or cytogenetics.22, 23, 24 Gastric MALT lymphomas have been reported to feature the highest incidence of t(11;18)(q21;q21), with a frequency ranging from 17–48% depending upon the kind of study.7, 8, 24, 25, 26, 27 In the present study, most t(11;18)(q32;q21) were detected in MALT lymphomas located within the gastrointestinal tract; of the 25 gastrointestinal MALT lymphomas, six cases (24%) harboured a t(11;18)(q32;q21). Only two t(11;18)(q32;q21)-positive cases arose at other sites, being the lung and the parotid gland. The presence of the t(1;14)(p22;q32) was demonstrated in only one case (1%). The rarity of the latter genetic aberrance makes it very difficult to properly comment its anatomic site of preference, but lung, stomach, parotid gland and skin were described in sporadic case reports and some larger studies.2, 23, 24, 28, 29 The t(1;14)(p22;q32)-positive MALT lymphoma in our study was located in the nasopharyngeal sinus and to the best of our knowledge, this represents the first case arising at this site. Two of the 77 MALT lymphomas were characterised by t(14;18)(q32;q21), corresponding to an overall incidence of 3%. For a reason not clear to us, this is a much lower incidence as compared to the data published by Streubel et al13, 23, 30 who documented the presence of t(14;18)(q32;q21) in 10% to 18% of all MALT lymphomas. Both t(14;18)(q32;q21)-positive MALT lymphomas in our series arose within the lachrymal gland, in line with reports that t(14;18)(q32;q21) is occurring in MALT lymphomas outside the gastrointestinal tract.13, 23 In a previous study by our group performed on the same series of MALT lymphomas,17 it was shown that only one case (1%), located in the stomach, displayed the recently described t(3;14)(p13;q32), in contrast to the high incidence (10%) and exclusively nongastric locations of FOXP1 gene rearrangements in MALT lymphomas as reported in the study by Streubel et al.31 In analogy with t(14;18)(q32;q21), we cannot explain this striking difference in incidence but find these data worthwhile to report since it provides complimentary information to the study by Streubel et al about the incidence of t(14;18)(q32;q21) and t(3;14)(p13;q32) in MALT lymphomas.
Besides structural chromosomal abnormalities, 16 cases (21%) harboured the presence of three copies of the MALT1 gene, most likely due to trisomy 18 (confirmed by cytogenetics in eight out of 35 cases where cytogenetics were available), a genetic feature well known and described in nodal, extranodal and splenic marginal zone lymphomas.28, 32, 33 Polysomy 18 was demonstrated in two cases including one with cytogenetically documented three copies of isochromosome 18q. MALT lymphomas with trisomy or polysomy 18 were mainly detected outside the gastrointestinal tract. One gastric MALT lymphoma was shown to have an amplification of the MALT1 gene, confirming the previous CGH results on this particular case.14 In total, 28 of the 77 MALT lymphomas (26%) harboured an abnormality of the MALT1 gene, being either a structural abnormality (t(11;18)(21;q21) or t(14;18)(q32;q21)) or a numerical aberrance (trisomy or polysomy 18, or an amplification).
Of interest, the four aberrant BCL10 staining patterns found by immunohistochemistry corresponded well with the genetic findings. All eight t(11;18)(q21;q21)-positive MALT lymphomas (100%) displayed a nuclear presence of the BCL10 protein (with or without cytoplasmic BCL10 expression), suggesting that this altered subcellular BCL10 localisation is linked to the pathway induced by the API2-MALT1 fusion. However, the case with the most abundant nuclear BCL10 expression did not harbour a t(11;18)(q21;q21) but a t(1;14)(p22;q21), implying that a control of the BCL10 gene by the Ig enhancer does not only result in a deregulated BCL10 expression but also in an altered subcellular localisation of the protein as previously described.5 Surprisingly, the two orbital MALT lymphomas with a t(14;18)(q32;q21) featured an unique immunohistochemical staining pattern, with the BCL10 protein being located and concentrated in a perinuclear area. A similar but less prominent perinuclear BCL10 localisation (in combination with nuclear BCL10 staining) was observed in the one gastric MALT lymphoma case displaying an amplification of the MALT1 gene without MALT1 rearrangement. We speculate that both t(14;18)(q32;q21) and MALT1 amplification lead to an increased cellular expression of the MALT1 protein, resulting in a perinuclear accumulation of the BCL10 protein. Although 10 out of the 18 cases with trisomy or polysomy 18 showed an aberrant BCL10 expression, no correlation with a particular pattern was found. Of interest, while all t(11;18)(q21;q21)- and t(1;14)(p22;q32)-positive cases displayed nuclear (with or without cytoplasmic) BCL10 staining, the reverse cannot be said as nuclear BCL10 positivity was also observed in part of the cases with tri- or polysomy 18 and cases without detectable BCL10 and MALT1 aberrations. Finally, MALT lymphomas with no or weak cytoplasmic BCL10 expression featured none of the analysed BCL10 or MALT1 rearrangements.
Except for the one case with a t(1;14)(p22;q32), our findings demonstrate that neither rearrangements, nor numeric aberrances, nor genomic mutations5 of the BCL10 gene play an important role in the abnormal subcellular localisation of the BCL10 protein associated with this malignancy. In contrast, our study supports the close interaction between the MALT1 and BCL10 protein in MALT lymphomas, as there is a clear correlation between the BCL10 staining pattern and the type of MALT1 rearrangement. Both BCL10 and MALT1 play a crucial role in the antigen receptor-signalling pathway leading to activation of NF-κB.34, 35, 36 In vitro and in vivo experiments have shown that MALT1 physically associates with BCL10: this interaction involves the two Ig-like domains of MALT1 and a short stretch of amino acids that follow the CARD motif of BCL10.37 In addition, it was demonstrated that MALT1 is synergistic with BCL10 to enhance NF-κB activation in both B and T cells, and that the oligomerisation and activation of MALT1 depends on BCL10. The latter is a possible explanation for the perinuclear localisation of BCL10 in MALT lymphomas with t(14;18)(q32;q21) or MALT1 amplification, as the abundantly expressed MALT1 might interact with BCL10 and stabilises it in the (perinuclear) cytoplasm, consequently leading to its accumulation.
Several lines of evidence support the hypothesis that MALT1 acts at the level of or downstream of BCL10 in the NF-κB-signalling pathway. In 2001, three groups independently identified CARMA1 (also known as CARD11 or BIMP1) as the upstream activator of BCL10;38, 39, 40 it binds BCL10 through a CARD–CARD interaction hereby forming a ternary complex with both BCL10 and MALT1. The group of Gaide et al demonstrated that overexpression of BCL10 in vitro resulted in a diffuse cytoplasmic BCL10 staining pattern while CARMA1 showed a granular perinuclear staining; however, upon coexpression with CARMA1, most of the cytoplasmic staining of BCL10 disappeared and BCL10 now colocalised with CARMA1 to perinuclear structures. Interestingly, in our series, t(14;18)(q32;q21)-positive MALT lymphomas as well as the MALT lymphoma with MALT1 amplification were marked by a similar perinuclear localisation of the BCL10 protein. As such, it may be that not only the downstream component of MALT1 plays a role in the subcellular localisation of BCL10, but also the upstream protein of CARMA1. However, in view of the low number of cases with this perinuclear BCL10 pattern and the lack of actual localisation data on CARMA1 in our series (properly working antibodies are not commercially available yet), this remains very hypothetical.
Finally, presence of aberrant BCL10 expression found by immunohistochemistry is helpful in the diagnosis of MALT lymphomas and this finding might be an indication of an underlying genetic anomaly either involving the BCL10 or MALT1 gene.
References
Jaffe E, Harris N, Stein H, et al. World Health Organisation Classification of Tumours: Pathology and Genetics: Tumours of Haemopoietic and Lymphoid Tissues. IAR Press: Lyon, France, 2004.
Willis TG, Jadayel DM, Du MQ, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 1999;96:35–45.
Thome M . CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol 2004;4:348–359.
Ye HT, Dogan A, Karran L, et al. BCL10 expression in normal and neoplastic lymphoid tissue—nuclear localization in MALT lymphoma. Am J Pathol 2000;157:1147–1154.
Maes B, Demunter A, Peeters B, et al. BCL10 mutation does not represent an important pathogenic mechanism in gastric MALT-type lymphoma, and the presence of the API2-MLT fusion is associated with aberrant nuclear BCL10 expression. Blood 2002;99:1398–1404.
Yeh K, Kuo S, Chen L, et al. Nuclear expression of BCL10 or nuclear factor kappa B helps predict Helicobacter pylori-independent status of low-grade mucosa-associated lymphoid tissue lymphomas with or without t(11;18)(q21;q21). Blood 2005;106:1037–1041.
Baens M, Steyls A, Geboes K, et al. The product of the t(11;18), an API2-MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol 2000;156:1433–1439.
Dierlamm J, Baens M, Stefanova-Ouzounova M, et al. Detection of t(11;18)(q21;q21) by interphase fluorescence in situ hybridization using API2 and MLT specific probes. Blood 2000;96:2215–2218.
Dierlamm J, Baens M, Wlodarska I, et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood 1999;93:3601–3609.
Starostik P, Patzner J, Greiner A, et al. Gastric marginal zone B-cell lymphomas of MALT type develop along 2 distinct pathogenetic pathways. Blood 2002;99:3–9.
Muller-Hermelink HK . Genetic and molecular genetic studies in the diagnosis of B-cell lymphomas: marginal zone lymphomas. Hum Pathol 2003;34:336–340.
Sanchez-Izquierdo D, Buchonnet G, Siebert R, et al. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood 2003;101:4539–4546.
Streubel B, Lamprecht A, Dierlamm J, et al. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood 2003;101:2335–2339.
Dierlamm J, Rosenberg C, Stul M, et al. Characteristic pattern of chromosomal gains and losses in marginal zone B cell lymphoma detected by comparative genomic hybridization. Leukemia 1997;11:747–758.
Dierlamm J, Pittaluga S, Stul M, et al. BCL6 gene rearrangements also occur in marginal zone B-cell lymphoma. Br J Haematol 1997;98:719–725.
Maes B, Baens M, Marynen P, et al. The product of the t(11;18), an API2-MLT fusion, is an almost exclusive finding in marginal zone cell lymphoma of extranodal MALT-type. Ann Oncol 2000;11:521–526.
Wlodarska I, Veyt E, De Paepe P, et al. FOXP1, a gene highly expressed in a subset of diffuse large B-cell lymphoma, is recurrently targeted by genomic aberrations. Leukemia 2005;19:1299–1305.
ISCN.In: Mitelman F (ed). An International System for Human Cytogenetic Nomenclature. Karger: Basel, 1995.
Dierlamm J, Wlodarska I, Michaux L, et al. Successful use of the same slide for consecutive fluorescence in situ hybridization experiments. Genes Chromosomes Cancer 1996;16:261–264.
Martin-Subero JI, Harder L, Gesk S, et al. Interphase FISH assays for the detection of translocations with breakpoints in immunoglobulin light chain loci. Int J Cancer 2002;98:470–474.
Poulsen TS, Silahtaroglu AN, Gisselo CG, et al. Detection of illegitimate rearrangement within the immunoglobulin locus on 14q32.3 in B-cell malignancies using end-sequenced probes. Genes Chromosomes Cancer 2001;32:265–274.
Ott G, Katzenberger T, Greiner A, et al. The t(11;18)(q21;q21) chromosome translocation is a frequent and specific aberration in low-grade but not high-grade malignant non-Hodgkin's lymphomas of the mucosa-associated lymphoid tissue (MALT-) type. Cancer Res 1997;57:3944–3948.
Streubel B, Simonitsch-Klupp I, Mullauer L, et al. Variable frequencies of MALT lymphoma-associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004;18:1722–1726.
Ye HT, Liu HX, Attygalle A, et al. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood 2003;102:1012–1018.
Nakamura S, Matsumoto T, Nakamura S, et al. Chromosomal translocation t(11;18)(q21;q21) in gastrointestinal mucosa associated lymphoid tissue lymphoma. J Clin Pathol 2003;56:36–42.
Remstein ED, James CD, Kurtin PJ . Incidence and subtype specificity of AP12-MALT1 fusion translocations in extranodal, nodal, and splenic marginal zone lymphomas. Am J Pathol 2000;156:1183–1188.
Yonezumi M, Suzuki R, Suzuki H, et al. Detection of AP12-MALT1 chimaeric gene in extranodal and nodal marginal zone B-cell lymphoma by reverse transcription polymerase chain reaction (PCR) and genomic long and accurate PCR analyses. Br J Haematol 2001;115:588–594.
Wotherspoon AC, Pan LX, Diss TC, et al. Cytogenetic Study of B-Cell Lymphoma of Mucosa-Associated Lymphoid-Tissue. Cancer Genet Cytogenet 1992;58:35–38.
Zhang QG, Siebert R, Yan MH, et al. Inactivating mutations and overexpression of BCL10, a caspase recruitment domain-containing gene, in MALT lymphoma with t(1;14)(p22;q32). Nat Genet 1999;22:63–68.
Streubel B, Huber D, Wohrer S, et al. Frequency of chromosomal aberrations involving MALT1 in mucosa-associated lymphoid tissue lymphoma in patients with Sjogren's syndrome. Clin Cancer Res 2004;10:476–480.
Streubel B, Vinatzer U, Lamprecht A, et al. T(3;4)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia 2005;19:652–658.
Dierlamm J, Pittaluga S, Wlodarska I, et al. Marginal zone B-cell lymphomas of different sites share similar cytogenetic and morphologic features. Blood 1996;87:299–307.
Offit K, Jhanwar SC, Ladanyi M, et al. Cytogenetic analysis of 434 consecutively ascertained specimens of non-Hodgkins-lymphoma—correlations between recurrent aberrations, histology, and exposure to cytotoxic treatment. Genes Chromosomes Cancer 1991;3:189–201.
Ruland J, Duncan GS, Elia A, et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 2001;104:33–42.
Ruland J, Duncan GS, Wakeham A, et al. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 2003;19:749–758.
Ruefli-Brasse AA, French DM, Dixit VM . Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science 2003;302:1581–1584.
Lucas PC, Yonezumi M, Inohara N, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem 2001;276:19012–19019.
Gaide O, Martinon F, Micheau O, et al. Carma1, a CARD-containing binding partner of Bcl10, induces Bcl10 phosphorylation and NF-kappa B activation (FEBS 24842) (vol 496, pg 121, 2001). FEBS Lett 2001;505:198.
Bertin J, Wang L, Guo Y, et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem 2001;276:11877–11882.
McAllister-Lucas LM, Inohara N, Lucas PC, et al. Bimp1, a MAGUK family member linking protein kinase C activation to Bcl10-mediated NF-kappa B induction. J Biol Chem 2001;276:30589–30597.
Acknowledgements
We thank Ursula Pluys and Lore Bernar for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sagaert, X., Laurent, M., Baens, M. et al. MALT1 and BCL10 aberrations in MALT lymphomas and their effect on the expression of BCL10 in the tumour cells. Mod Pathol 19, 225–232 (2006). https://doi.org/10.1038/modpathol.3800523
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.3800523
Keywords
This article is cited by
-
Primary MALT lymphoma of the breast: pathological and radiological characteristics
Breast Cancer Research and Treatment (2024)
-
Molecular pathogenesis of lymphomas of mucosa-associated lymphoid tissue—from (auto)antigen driven selection to the activation of NF-κB signaling
Science China Life Sciences (2015)
-
T(14;18)(q32;q21) involving MALT1 and IGH genes occurs in extranodal diffuse large B-cell lymphomas of the breast and testis
Modern Pathology (2013)
-
Gastric MALT lymphoma: a model of chronic inflammation-induced tumor development
Nature Reviews Gastroenterology & Hepatology (2010)
-
Comparative expressed sequence hybridization studies of t(11;18)(q21;q21)-positive and -negative gastric MALT lymphomas reveal both unique and overlapping gene programs
Modern Pathology (2010)