Abstract
Melanoma inhibitory activity (MIA) has been identified as a small protein secreted from malignant melanoma cells. Recent results revealed a direct interaction of MIA and epitopes within extracellular matrix proteins including fibronectin. The aim of this study was to analyze functional consequences mediated by this interaction. Here we show that MIA interferes specifically with attachment of melanoma cells to fibronectin, a phenomenon we refer to as active detachment. Antibodies inhibiting binding of α4β1 and α5β1 integrins to fibronectin cross-react specifically with MIA, suggesting that MIA shares significant structural homology with the binding pockets of these integrins and thereby masks the respective epitopes on extracellular matrix molecules. Several peptides derived from fibronectin and from a phage display screening were tested with respect to a potential MIA-inhibitory effect. In vitro tests identified two peptides affecting MIA function; both inhibited growth of melanoma metastases in vivo. In summary, we conclude that MIA may play a role in tumor progression and spread of malignant melanomas via mediating active detachment of cells from extracellular matrix molecules within their local milieu. Further, our results suggest that inhibiting MIA functions in vivo may provide a novel therapeutic strategy for metastatic melanoma disease.
Similar content being viewed by others
Main
We have previously identified melanoma inhibitory activity (MIA), an 11-kd protein secreted from malignant melanoma cells into the tissue culture supernatant (Apfel et al, 1992; Blesch et al, 1994; Bogdahn et al, 1989). Comparison of the primary peptide sequence revealed that MIA represents a novel protein without any homology to previously known protein families or protein domains. Purified MIA induces growth inhibition of malignant melanoma cells in vitro and causes significant alteration of cell morphology as melanoma cells round up (Blesch et al, 1994). Despite its growth-inhibitory effect on melanoma cell lines in vitro, MIA expression in vivo correlates with progressive malignancy of melanocytic tumors (Bosserhoff et al, 1996; 1999). In recent studies we have been able to measure enhanced MIA protein levels specifically in the serum of patients with metastatic melanomas (Bosserhoff et al, 1997; Dreau et al, 1999). Analyzing normal skin and skin-derived melanocytic tumors by semiquantitative RT-PCR did not reveal significant MIA mRNA levels in normal skin and melanocytes but moderate levels in the majority of benign melanocytic nevi and very high levels in all primary and metastatic malignant melanomas (Bosserhoff et al, 1996; 1999). Absence of MIA mRNA expression in benign melanocytes cultured from normal skin biopsies has also been reported by a different study (van Groningen et al, 1995). We have further sequenced MIA cDNA clones from different sources, including cDNAs from several melanoma cell lines and genomic clones from human placental DNA, and confirmed that the MIA gene is not mutated in malignant melanomas. Additionally, in vivo studies suggested the importance of MIA in melanoma metastasis (Bosserhoff et al, 2001; Guba et al, 2000). In summary, these data do not support the idea of MIA acting as a tumor suppressor gene in vivo.
Recently we have analyzed the three-dimensional structure of MIA by multidimensional nuclear magnetic resonance (NMR) and have shown that the peptide adopts an SH3 domain-like fold in solution and directly interacts with fibronectin (Stoll et al, 2001). Based on these results and on the observation that MIA alters melanoma cell morphology, we decided therefore to investigate the possibility that the growth-inhibitory effect in vitro results from interference with cell attachment by decreasing interactions between melanoma cells and extracellular matrix (ECM) molecules.
Results
Co-immunoprecipitation of MIA and Fibronectin
We have previously shown that MIA adopts an SH3 domain-like structure in solution and directly interacts with fibronectin and peptides derived from fibronectin (Stoll et al, 2001). To support this data further, co-immunoprecipitation experiments were performed, which indicated specific interaction between fibronectin and MIA but not between MIA and collagen type I (Fig. 1). The immunoprecipitated proteins were analyzed by Western blot and were immunoprobed with a polyclonal rabbit MIA antiserum. The results are consistent with data obtained from previous MIA-peptide interaction studies and indicate that MIA does not stick to ECM molecules due to unspecific protein interaction but rather binds specifically to a subset of epitopes, particularly fibronectin. Previous data also revealed binding of MIA to different peptides derived from Fn6, Fn10, and Fn14 domains (Stoll et al, 2001). Further, co-immunoprecipitation showed interaction between MIA and laminin.
Co-immunoprecipitation of melanoma inhibitory activity (MIA) and fibronectin. One μg of recombinant MIA was added to 1 μg of fibronectin (Fn), laminin (Ln), or collagen type I (Coll I) and immunoprecipitated with an antibody directed against the respective extracellular matrix (ECM) molecule. The immunoprecipitate was then separated on a SDS-PAGE gel, Western blotted, and immunoprobed for MIA. MIA was detected when co-precipitated with laminin (Lane 1) and fibronectin (Lane 2) but not with collagen type I (Lane 3). None of the antibodies directed against fibronectin or collagen type I precipitated MIA in the absence of the respective ECM molecule (Lanes 5 to 7). Bands at 50 kd and 25 kd are representing the heavy and the light chain of IgG.
Specific Interactions between MIA and Peptide Epitopes in Fibronectin
Different ECM molecules or internal peptide epitopes were coated onto a plastic surface and exposed to MIA. The amount of MIA that was bound to the coated ECM molecule was measured by a peroxidase-linked monoclonal MIA antibody. Results shown in Tables 1 and 2 reveal that MIA binds specifically to surfaces coated with fibronectin, laminin, and tenascin; and further to surfaces coated with peptides CS1 (connecting segment), CS5, the fibronectin-like engineered protein (multimerized arginine-glycine-aspartate [RGD]-peptide epitopes) (Esty, 1991); and fibronectin-derived peptides FN6, Fn10, and Fn14. In contrast, MIA did not bind to collagen types I and II, heparan sulfate proteoglycan (HSPG), vitronectin, laminin-derived peptides, or other control peptides, and only weakly to collagen type IV. To provide further evidence that the binding is specific and not due to the “stickiness” of ECM proteins, the binding assays were performed with denatured MIA protein. Denatured MIA protein is still detected by the polyclonal antibody used in the assay, but the three-dimensional structure requiring stabilization by two disulfide bonds is destroyed. Denatured MIA was not able to bind to any of the matrix proteins tested (data not shown).
Cross-reaction of Anti-Integrin Antibodies and MIA
As pointed out above, all peptides and proteins that bind to MIA serve as binding sites for certain integrins (Hemler et al, 1987; Mohri et al, 1996; Mould et al, 1991), raising the possibility that MIA and the ligand binding pockets of these integrins share three-dimensional homology. Addressing this possibility we asked whether monoclonal antibodies that inactivate the binding pockets of integrins (Hemler et al, 1987) cross-react with MIA. In parallel, controls were performed with anti-integrin antibodies recognizing epitopes outside the binding pocket (Bergelson et al, 1992; Falcioni et al, 1986; Teixido et al, 1992) and with antibodies recognizing cell surface epitopes other than integrins (E-Cad, EGF-Rec, and HLA-DR). Positive control reactions were performed with two monoclonal anti-MIA antibodies. Results from these immunoreactions (Fig. 2A) revealed that the four antibodies that inactivate the binding pockets of α4 and α5 integrins cross-react specifically with MIA in contrast to all other integrin and cell surface molecule antibodies. These results strongly suggest that the activated binding pockets of α4β1 and α5β1 integrin receptors share significant three-dimensional homology with MIA. In addition, a sequence alignment using ClustalW software (EBI, Cambridge, United Kingdom) further corroborates these findings. A low yet detectable homology between human integrin α4, α5, and human MIA is found for the β-propeller domain of integrins (Fig. 2B).
Specific cross-reaction of anti–α4 and -α5-integrin–inhibiting antibodies with melanoma inhibitory activity (MIA). (A) Bacterially purified MIA was coated to 96-well plates and incubated for 30 minutes with 1 μg/ml of the following monoclonal antibodies: α2-, α3-, α4-, α5-, α6-, αv-, and β1-integrin inhibitory (i) antibodies α2-, α4-, α5-, α6-, αv- and β1-integrin noninhibitory (ni) antibodies HLA-DR, E-cadherin, epidermal-growth-factor receptor, or β-galactosidase antibodies and two different MIA antibodies. After washing five times with PBS BSA, binding of the respective monoclonal antibodies was detected by a horseradish peroxidase-coupled antimouse IgG antiserum, and then visualized with ABTS (Roche) and quantified after 30 minutes at OD 405nm. (B) Sequence alignment of integrin α4, integrin α5, and human MIA. The Swiss-Prot primary accession numbers are P13612 for human integrin-α4, P08648 for human integrin α5, and Q16674 for human MIA. The color coding is red for identical residues in all of the three sequences (*), green for the strongly similar residues (:), and blue for weakly similar residues (.). The primary consensus sequence is also shown. For further details, please refer to the “Cross-reaction of Anti-Integrin Antibodies and MIA” section.
To demonstrate the importance of the three-dimensional structure and correct folding of the MIA protein, the same assay was repeated using reduced MIA protein. As it is known that the two cystein bonds are essential for MIA folding, reducing conditions lead to denaturing of the protein and to a reduced solubility as monitored by NMR spectroscopy (data not shown). All antibodies that cross-reacted with native MIA were incapable of binding to denatured MIA protein (data not shown). Further, Western blot analysis was performed using the same panel of anti-integrin antibodies. Again, none of the antibodies detecting native MIA protein reacted with MIA under reducing conditions (data not shown).
MIA Regulates Attachment of Melanoma Cells
To test the functional relevance of MIA secretion by melanoma cells, the effect of MIA on adhesion and invasion was analyzed. The ability of melanoma cells to adhere to different ECM proteins was measured in the presence or absence of MIA. Because recombinant human MIA protein was used for these analyses, we studied the human melanoma cell line Mel Im in parallel to murine B16 cells. Addition of 50 ng/ml MIA reduced attachment of Mel Im cells to fibronectin, laminin, and tenascin between 30% and 50% in parallel to a significant change in cell morphology (Fig. 3, A and B). In the presence of MIA a large portion of cells attaching to fibronectin and laminin but not to HSPG acquired a rounded-up, lymphoid-like cell morphology. Consistently, adhesion to matrigel representing a reconstituted basement membrane rich in laminin, fibronectin, and tenascin was also inhibited. This effect was dose-dependent, with a maximal inhibition of 50% at 50 ng/ml (data not shown) and was also observed with every other human melanoma cell line (Mel Wei, Mel Ju, Mel Ho, Mel Ei, Mel Juso, and HTZ-19d) that we tested. In contrast, addition of MIA did not alter significantly adhesion of Mel Im cells to collagen types I, II, IV, vitronectin, and HSPG (Fig. 3B). Identical results were obtained for the murine B16 melanoma cell line with respect to reduced adhesion to fibronectin and tenascin (data not shown). As MIA did not influence chemotaxis measured in a Boyden chamber model or secretion of matrix metalloproteases from melanoma cells (data not shown), we were able to confirm the results from attachment analyses in Boyden chamber invasion assays. In this assay melanoma cells are placed into the upper chamber, which is separated from a second compartment by a filter coated with different ECM molecules. The number of cells that attach to and invade through the filters can be quantified reliably (Albini et al, 1987; Terranova et al, 1986). The results from both Boyden chambers and attachment assays parallel closely and reveal consistently that MIA inhibits interaction of melanoma cells with fibronectin, laminin, tenascin, and matrigel (Fig. 3C) and therefore inhibits invasion in this in vitro model. Furthermore, our findings provide an explanation of how MIA exerts its growth inhibitory effects on cultured melanoma cells in vitro. To analyze if MIA also has an effect on other cell types, we performed proliferation and migration assays with fibroblasts, keratinocytes, endothelial cells, and lymphocytes. No differences in proliferation and in the migratory potential of the analyzed cell types were seen (data not shown).
Inhibition of melanoma cell attachment to matrix proteins by melanoma inhibitory activity (MIA). (A) Mel Im melanoma cells were seeded onto plastic surfaces coated with fibronectin (Fn), laminin (Lm), and heparan sulfate proteoglycan (HSPG) and grown for 6 hours. In the presence of 50 ng/ml bacterially purified MIA (3) cells round up and appear significantly less adhesive to Fn and Lm. (B) Adhesion of Mel Im melanoma cells to 96-well plates coated with the following matrix proteins: fibronectin (1 μg/cm2), matrigel (1:3 diluted in H2O), laminin (5 μg/cm2), tenascin (1 μg/cm2), collagen types I, II, and IV (1 μg/cm2), vitronectin (50 ng/cm2), or HSPG (2 μg/cm2). **p < 0.001; *p < 0.005. (C) Boyden chamber assays quantifying adhesion and invasion of Mel Im melanoma cells through matrix proteins. Polycarbonate filters with 8-μm pore size were coated with extracellular matrix molecules as described in B. Fibroblast-conditioned medium was placed as a chemoattractant into the lower compartment. After incubation at 37° C for 4 hours, cells adhering to the lower filter surface were fixed, stained, and counted. **p < 0.001; *p < 0.005.
Specific Peptide Epitopes Present in Fibronectin Antagonize the Effect of MIA on Attachment
Attachment of melanoma cells to ECM involves specific interactions between cell surface integrin receptors and epitopes in ECM molecules including fibronectin. Results shown in Figure 3B underline further that fibronectin is an ECM molecule promoting strong adhesion of melanoma cells. Therefore, we tested whether peptides derived from epitopes in fibronectin identified by previous experiments as interacting with MIA were able to inhibit MIA function. Two peptides, CS1 (EILDVPST) and CS5 (RQDV), serving as α4β1 and α5β1 integrin binding epitopes within the type III connecting segment (III CS) of fibronectin (Humphries et al, 1986; Mould et al, 1991), reduced significantly the inhibitory effect of MIA (Table 3). Consistently, the entire fibronectin molecule, a large protein composed of multimerized RGD sites (fibronectin-like engineered protein) and the three peptides Fn6, Fn10, and Fn14 were also able to antagonize MIA. In contrast, other peptide epitopes that mediate adhesion independent of α4β1 or α5β1 integrins, fibronectin adhesion promoting peptide (WQPPRARI) (Woods et al, 1993), laminin chain α peptide (SIKVAV, targeted by α1β1 and αvβ3 integrins), and YIGSR-peptide (targeted by the 67-kd laminin receptor) did not inhibit the effect of MIA.
Effect of MIA-Inhibitory Peptides on Growth of Melanoma Metastasis In Vivo
Both in vitro and in vivo MIA expression levels strictly correlate with a highly invasive phenotype (Bosserhoff et al, 1996, 1999; 2000). Further in vivo studies have shown the necessity of MIA for melanoma invasion and metastasis (Bosserhoff et al, 2001; Guba et al, 2000). We therefore aimed to design MIA-inhibitory peptides based on our experiments that identified fibronectin epitopes interacting with MIA ((Stoll et al, 2001) (Table 2). Peptides derived from fibronectin, which were shown previously to cross-react with MIA, were tested in Boyden chamber assays on their effect on melanoma cell invasion with and without MIA. The results shown in Table 3 clearly reveal that the peptides Fn6, Fn10, and Fn14 block MIA function and further that Fn10 and Fn14 also inhibit tumor cell attachment. We dismissed Fn10 and Fn14 from further in vivo testing because we expected them also to influence attachment of other cell types. In addition, we tested several of the peptide sequences derived from phage display screenings recently published (Stoll et al, 2001) and found peptide GTQSTAIPAPTD (peptide 878) to be a potentially interesting candidate for therapeutic MIA inhibition. It has a high homology to the fibronectin type III module 12/13, and functional analysis showed no effect on melanoma cell attachment but strongly inhibited MIA function. Both peptides, Fn6 and 878, were analyzed for effects on proliferation or apoptosis of the melanoma cells. No differences comparing peptide-treated and untreated cells were observed (proliferation: doubling time 22.2 hours [untreated] versus 21.6 hours [peptide Fn6] or 21.9 hours [peptide 878]).
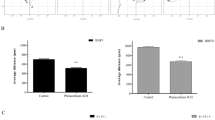
We then tested the effect of peptide Fn6 and peptide 878 on the growth of metastasis in vivo using the B16/C57Bl6 model (Fig. 4). The number of black lung nodules of melanoma was not significantly changed between treated and untreated controls: 193 ± 13 (control), 249 ± 26 (Fn6), and 198 ± 19 (peptide 878). However, significant changes were seen in the size of the nodules. Animals treated with the peptides developed significantly smaller tumor nodules compared with the control animals: 119.9 mm2 ± 9.95 (control); 87.15 mm2 ± 5.32, p = 0.0042 (Fn6); 78.1 mm2 ± 5.03, p = 0.0003 (peptide 878). The tumors were analyzed histologically and immunohistologically (Fig. 5). Hematoxylin and eosin staining did not reveal morphologic differences (Fig. 5A). Further, no changes in proliferation were determined counting the rate of mitosis in the tumor. In all tumors small foci of necrosis and single lymphocyte infiltration were found in equal number. To verify melanocytic origin of the cells, the tumors were stained with HMB45 (Fig. 5B). Angiogenesis in the tumor was visualized by staining factor VIII (Fig. 5C). No difference in the amount or distribution of vessels was seen comparing treated and untreated animals.
Effect of melanoma inhibitory activity (MIA)-inhibitory peptides on growth of melanoma metastasis in vivo. (A) The number of black lung modules of melanoma was not significantly changed comparing untreated and treated animals (ns = not significant). (B) The size of black lung modules of melanoma was significantly changed after treatment with MIA-inhibitory peptides (p values are given).
Effect of melanoma inhibitory activity (MIA)-inhibitory peptides on melanoma metastasis in vivo. (A) Tumors of treated and untreated mice were analyzed histologically. No changes in morphology, lymphocyte infiltration, or necrosis could be seen. Counting of mitosis to evaluate rate of proliferation (arrows) revealed no differences comparing treated and untreated animals. (B) To verify melanocytic origin of the tumors, HMB45 staining was performed. (C) The tumors were stained with an anti–factor VIII antibody to analyze vascularization of tumor tissue. No differences comparing treated and untreated animals were determined.
Discussion
Experimental data described in this study provide strong evidence for specific interactions between MIA and the ECM protein fibronectin and suggest that secretion of MIA from melanoma cells results in masking of integrin binding sites within the local environment. Because previous studies (Bosserhoff et al, 2001; Guba et al, 2000) demonstrated tight correlation between MIA expression levels and the metastatic phenotype of melanoma cells, we speculate that MIA enables melanoma cells to detach from some of their ECM contacts. Consequently, adhesion of melanoma cells to fibronectin, laminin, and tenascin is reduced by 30% to 50%. Taken together, our data identify active detachment through secretion of MIA as a novel molecular mechanism by which neoplastic melanocytes specifically change their attachment to components of the ECM and basement membranes to enhance their metastatic capability. Interactions between cell surface molecules and matrix components do not only mediate physical adhesion but also provide important signals for melanocytic cells. Therefore, detachment of melanoma cells by MIA may also be implicated in regulation of migration, apoptosis, secretion of proteases or matrix proteins, and cell growth (Bates et al, 1995; Boudreau et al, 1995; Clarke et al, 1995; Langholz et al, 1995; Varner et al, 1995). It is known that such interactions between melanocytic cells and ECM involve foremost binding of integrins to specific epitopes within fibronectin and depend, to a significant extent, on activation of α4β1 and α5β1 integrins (Scott et al, 1992). Detachment from ECM molecules within the local milieu is a key prerequisite for melanoma cells to migrate, invade, and finally metastasize as a systemic disease. A previous study has demonstrated that overexpression of functionally active α4β1 integrin in B16 cells reduced strongly matrigel invasion and pulmonary metastasis (Qian et al, 1994). Therefore, MIA particularly affects interactions between ECM molecules and integrins, which play critical roles in controlling melanoma cell metastases at the invasive stage. Active detachment promoted by MIA secretion appears to be a highly regulated and cell-type–specific mechanism because other cell types known to migrate actively including macrophages or lymphocytes do not express MIA based on analyses by RT-PCR or ELISA.
Interestingly a similar mechanism involving secretion of a molecule modulating cell-matrix contacts and invasion of melanoma cells was described previously for SPARC and NG2 (Burg et al, 1998; Ledda et al, 1997). Therefore, we conclude that secretion of MIA from melanoma cells below the amount required for growth inhibition in vitro results in masking of important attachment sites within the local environment and takes part in a large amount of concerted changes in tumor cell-matrix interaction. This control of cell adhesion plays an important role in control of migration as we recently could show (Bosserhoff et al, 2001; Guba et al, 2000). Reducing MIA expression in melanoma cell lines results in less migratory ability of the cells, whereas an overexpression of MIA leads to stronger migratory activity.
We have recently determined the three-dimensional structure of MIA by NMR (Stoll et al, 2001), and our data were subsequently confirmed by X-ray analysis (Lougheed et al, 2001). It was shown that MIA forms a small globular SH3 domain-like structure stabilized by two disulfide bonds. By sequence comparison of MIA and α4 or α5 integrin weak homology in amino acid identity (18% or 15.6%) and homology in predicted secondary structure by hydrophobicity blots was detected (data not shown). Homology was found in the region of the W6 and W7 β-propeller structure in α4 and α5 integrin, a region known to be involved in ligand binding (Loftus et al, 1994; Ruoslahti, 1996; Springer, 1997).
In addition, a sequence alignment using ClustalW software (EBI) further corroborates these findings. A low yet detectable homology between human integrin α4, α5, and human MIA is found for the β-propeller domain of integrins (Fig. 2B). Interestingly, homology to integrin α4 and α5 residues is found throughout the entire sequence of human MIA. Because denatured human MIA does not cross-react with the anti-integrin α4 and α5, these antibodies obviously recognize a three-dimensional epitope in human MIA. However, a detailed analysis on the molecular level awaits the structure elucidation of integrin α4 and α5. This will allow the question as to which regions of human MIA are structurally similar to integrin α4 and α5 to be addressed.
Based on structural analyses, we were able to show that short peptides either derived from fibronectin or from a phage display screening are functional ligands of MIA and thus were tested functionally for MIA inhibition (Stoll et al, 2001). We were especially interested in testing peptides that interacted with MIA but did not significantly reduce cell adhesion. Two fibronectin-derived peptides (Fn6 and peptide 878) were identified as being able to inhibit MIA function in vitro without affecting cell adhesion and subsequently were tested in vivo for inhibition of metastasis in a mouse melanoma model. Application of both peptides led to a reduction in tumor size. We hypothesize that this is possibly due to less effective migration or invasion because in vitro experiments did not show an influence of the peptides used on melanoma cell proliferation or apoptosis. Further, data previously published point out that MIA is needed for migration and invasion of melanoma cells (Bosserhoff et al, 2001; Guba et al, 2000). Changes in the mode of application will now be tested in further studies to investigate a potential therapeutic application of MIA inhibitory peptides for metastatic disease. In summary, our data strongly suggest that inhibition of MIA function in vivo may provide a novel therapeutic strategy for therapy of patients with metastatic melanoma and provide the first proof of principle that the strategy is effective in an experimental metastasis model in vivo.
Materials and Methods
Origin of Cell Lines and Culture Conditions
The cell line HTZ-19d was established from a brain metastasis of a patient with a primary cutaneous malignant melanoma (Blesch et al, 1994). Melanoma cell lines Mel Im, Mel Wei, Mel Juso, Mel Ju, Mel Ho, and Mel Ei were established by Dr. M. Hadem (University of Hanover, Germany) and have been characterized previously (Grimm and Johnson, 1995; Jacob et al, 1995). The cell line SK-Mel 28 was obtained from ATCC (Rockville, Maryland). All cells were cultured in DMEM supplemented with 10% FCS and L-glutamine. Proliferation was measured using the XTT assay (Roche, Mannhein, Germany).
Expression and Purification of Recombinant MIA Protein
Escherichia coli M15(pREP4) cells transfected with the expression plasmid pQE40-MIA expressing 108 residues of human MIA (G25 to Q131) were grown to an absorbance OD600 nm = 0.6, induced by 1 mm isopropyl-1-thio-β-d-galactopyranoside for 4 hours and lysed by sonication. The protein was renatured from E. coli inclusion bodies as previously published (Jaenicke and Rudolph, 1986). Refolded human MIA was applied to hydrophobic interaction chromatography and further purified on a S-Sepharose Fast Flow (Sigma, St. Louis, Missouri). Finally, gel filtration was performed on a Superdex 200 Prep Grade (Sigma). Fractions containing human MIA were pooled and concentrated. Purified protein was checked by SDS-Page and HPLC and was shown to be 95% pure (data not shown).
Assays for Protein-Protein Interaction
ECM proteins listed under attachment assays or peptides derived from ECM molecules were coated onto 96-well plates and exposed to 50 ng/ml purified MIA (90 nm) for 30 minutes. MIA binding was quantified using a peroxidase-coupled monoclonal anti-MIA antibody and the substrate ABTS (Roche). Controls were exposed to 50 ng/ml BSA or to denatured MIA instead of native MIA. Reactions were quantified at an OD of 405 nm.
For co-immunoprecipitation, 1 μg of recombinant human MIA (90 nm) and 1 μg of the respective ECM molecule dissolved in PBS (fibronectin, 5 nm; laminin, 2 nm; and collagen type I, 2.6 nm) were precleared with 25 μl protein G-Sepharose (Pharmacia, Freiburg, Germany) at 4° C for 4 hours. After centrifugation the supernatant was transferred into a fresh vial and incubated with the respective antibody with vigorous shaking at 4° C overnight. Then 20 μl protein G-Sepharose was added for 1 hour, pelleted, washed three times with PBS, resuspended in Laemmli’s buffer, heated at 95° C for 10 minutes, and separated on 15% SDS-polyacrylamide gels. MIA was detected after Western blotting using the rabbit polyclonal MIA antiserum PAK 1881 (Roche).
MIA Immunoassay
MIA was coated onto 96-well plates and incubated for 30 minutes with 1 μg/ml of the following monoclonal antibodies: anti-α2 integrin (P1E6, Dako, Hamburg, Germany), anti-α2 integrin (A2–11E10, UBI, Lake Placid, New York) (Bergelson et al, 1992), anti-α3 integrin (P1B5, Dako), anti-α4 integrin (A4-PUJ1, UBI) (Hemler et al, 1987), anti-α4 integrin (B-5G10, UBI) (Teixido et al, 1992), anti-α4 integrin (P4C2, Chemicon), anti-α4 integrin (P1H4, Chemicon), anti-α4 integrin (AB1924, Chemicon), anti-α5 integrin (A5-PUJ5, UBI), anti-α5 integrin (P1D6, Chemicon), anti-α5 integrin (AB1949, Chemicon), anti-α6 integrin (A6-ELE, UBI) (Falcioni et al, 1986), anti-α6 integrin (provided by Dr. E. Klein, Würzburg, Germany), anti-αv integrin (P3G8, Chemicon), anti-αv integrin (AB1930, Chemicon), anti-αvβ3 integrin (Lv 230), anti-αvβ3 integrin (LM609, Chemicon), anti-β1 integrin (6S6, Chemicon), anti-β1 integrin (AB1952, Chemicon), anti–HLA-DR (CR3743, Dako, Hamburg, Germany), anti–E-cadherin (MLCA, EuroDiagnostica, Germany) and anti–EGF-receptor antibody (Clone F4, Sigma) and two different anti-MIA antibodies (Dr. B. Kaluza, Roche). After washing five times with PBS 3% BSA, binding of the respective mAb was detected by a horseradish peroxidase-coupled antimouse IgG antiserum, and then visualized with ABTS (Roche) and quantified at OD 405 nm after 30 minutes. Denatured MIA was used as a control for specificity in the same kind of assay.
Invasion and Attachment Assay
Invasion assays were performed in Boyden chambers containing polycarbonate filters with 8-μm pore size (Costar, Bodenheim, Germany) essentially as described previously (Albini et al, 1987; Terranova et al, 1986; Wach et al, 1996). Filters were coated with a commercially available reconstituted basement membrane (Matrigel, diluted 1:3 in H2O; Becton Dickinson #356235, Heidelberg, Germany), fibronectin (1 μg/cm2, 12.5 nmol/filter; Becton Dickinson #40008), laminin-1 (5 μg/cm2, 5 nmol/filter; Becton Dickinson #40232), tenascin (1 μg/cm2, 1 nmol/filter; Becton Dickinson #40239), collagen type I (1 μg/cm2, 6.5 nmol/filter; Becton Dickinson #40243), type II (1 μg/cm2, 6.5 nmol/filter; Becton Dickinson #40234), and type IV (1 μg/cm2, 5 nmol/filter; Becton Dickinson #40233), vitronectin (50 ng/cm2, 1 nmol/filter; Becton Dickinson #40238), or HSPG (2 μg/cm2, 1 nmol/filter; Becton Dickinson #40250). The lower compartment was filled with fibroblast-conditioned medium as a chemoattractant. Melanoma cells were harvested by trypsinization for 2 minutes, resuspended im DMEM without FCS at a density 2 × 105 cells/ml, and placed in the upper compartment of the chamber. Treatments were performed using MIA at a concentration of 50 ng/ml or RGD peptides at a concentration of 50 μg/ml. After incubation at 37° C for 4 hours, filters were removed. Cells adhering to the lower surface were fixed, stained, and counted.
Attachment assays were performed in 96-well plates. The wells were coated with the same matrix proteins as used for invasion assays by incubation at 4° C for 12 hours. Unspecific binding of cells was blocked by preincubation of the wells with 3% BSA PBS for 2 hours. Melanoma cells were harvested by trypsin incubation for 2 minutes, resuspended in DMEM without FCS at a density of 2 × 105 cells/ml with or without MIA (50 ng/ml), and placed in the respective wells. Cells remaining in suspension after 15 minutes were counted.
In Vivo Metastasis Assay
To measure the effect of peptides Fn6 (ETTIVITWTPAPR) and peptide 878 (GTQSTAIPAPTD) on the metastatic potential of B16 melanoma cells in vivo, an experimental metastasis assay was used (Bosserhoff et al, 2001; Qian et al, 1994). Iv injections of monodispersed tumor cells (4 × 105 cells per animal) were performed on syngeneic C57Bl6 mice (n = 8 for every peptide as well as control). Peptides (550 μg/mouse) or PBS was injected ip every day. After 16 days the mice were killed and the lungs were removed, washed in PBS, and fixed in formalin; the visible tumor nodules on the surface of the lungs were then counted, and the tumor areas measured. Statistical analyses were performed using the two-tailed Student’s t test. Paraffin-embedded sections of the tissues were histologically analyzed after hematoxylin and eosin staining, and stained for HMB45 and factor VIII expression by immunohistochemistry. Immunohistochemical studies for the expression of HMB45 (DAKO, 1:50) and factor VIII (DAKO, 1:600) were performed on tissue samples using an avidin-biotin peroxidase method with diaminobenzidine chromogen. After antigen retrieval (microwave treatment of formalin-fixed, paraffin-embedded 2- to 3-μm tissue sections for 40 minutes at 240 W in citrate buffer, pH 6.0), immunohistochemistry was carried out in a NEXES immunostainer (Ventana Medical System, Tucson, Arizona) according to the manufacturer’s instructions.
Sequence Analysis
Protein sequences were obtained from Swiss-Prot and analyzed using the software package ClustalW (EBI).
References
Albini A, Iwamoto Y, Kleinman HK, Martin GR, Aaronson SA, Kozlowski JM, and McEwan RN (1987). A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 47: 3239–3245.
Apfel R, Lottspeich F, Hoppe J, Behl C, Durr G, and Bogdahn U (1992). Purification and analysis of growth regulating proteins secreted by a human melanoma cell line. Melanoma Res 2: 327–336.
Bates RC, Lincz LF, and Burns GF (1995). Involvement of integrins in cell survival. Cancer Metastasis Rev 14: 191–203.
Bergelson JM, Shepley MP, Chan BM, Hemler ME, and Finberg RW (1992). Identification of the integrin VLA-2 as a receptor for echovirus 1 [see comments]. Science 255: 1718–1720.
Blesch A, Bosserhoff AK, Apfel R, Behl C, Hessdoerfer B, Schmitt A, Jachimczak P, Lottspeich F, Buettner R, and Bogdahn U (1994). Cloning of a novel malignant melanoma-derived growth-regulatory protein, MIA. Cancer Res 54: 5695–5701.
Bogdahn U, Apfel R, Hahn M, Gerlach M, Behl C, Hoppe J, and Martin R (1989). Autocrine tumor cell growth-inhibiting activities from human malignant melanoma. Cancer Res 49: 5358–5363.
Bosserhoff AK, Echtenacher B, Hein R, and Buettner R (2001). Functional role of melanoma inhibitory activity in regulating invasion and metastasis of malignant melanoma cells in vivo. Melanoma Res 11: 417–421.
Bosserhoff AK, Hauschild A, Hein R, Schadendorf D, Stockflesh E, Bogenrieder T, Landthaler M, Buettner R, and Stolz W (2000). Elevated MIA serum levels are of relevance for management of metastasized malignant melanomas: Results of a German multicenter study. J Invest Dermatol 114: 395–396.
Bosserhoff AK, Hein R, Bogdahn U, and Buettner R (1996). Structure and promoter analysis of the gene encoding the human melanoma-inhibiting protein MIA. J Biol Chem 271: 490–495.
Bosserhoff AK, Kaufmann M, Kaluza B, Bartke I, Zirngibl H, Hein R, Stolz W, and Buettner R (1997). Melanoma-inhibiting activity, a novel serum marker for progression of malignant melanoma. Cancer Res 57: 3149–3153.
Bosserhoff AK, Moser M, Hein R, Landthaler M, and Buettner R (1999) In situ expression patterns of melanoma-inhibiting activity (MIA) in malignant melanomas and breast cancers. J Pathol 187: 446–454.
Boudreau N, Sympson CJ, Werb Z, and Bissell MJ (1995). Suppression of ICE and apoptosis in mammary epithelial cells by extracellular matrix. Science 267: 891–893.
Burg MA, Grako KA, and Stallcup WB (1998). Expression of the NG2 proteoglycan enhances the growth and metastatic properties of melanoma cells. J Cell Physiol 177: 299–312.
Clarke AS, Lotz MM, Chao C, and Mercurio AM (1995). Activation of the p21 pathway of growth arrest and apoptosis by the beta 4 integrin cytoplasmic domain. J Biol Chem 270: 22673–22676.
Dreau D, Bosserhoff AK, White RL, Buettner R, and Holder WD (1999). Melanoma-inhibitory activity protein concentrations in blood of melanoma patients treated with immunotherapy. Oncol Res 11: 55–61.
Esty A (1991). Receptor-specific serum-free cell attachment using a highly stable engineered protein polymer. Am Biotechnol Lab 9: 44.
Falcioni R, Kennel SJ, Giacomini P, Zupi G, and Sacchi A (1986). Expression of tumor antigen correlated with metastatic potential of Lewis lung carcinoma and B16 melanoma clones in mice. Cancer Res 46: 5772–5778.
Grimm T and Johnson JP (1995). Ectopic expression of carcinoembryonic antigen by a melanoma cell leads to changes in the transcription of two additional cell adhesion molecules. Cancer Res 55: 3254–3257.
Guba M, Bosserhoff AK, Steinbauer M, Abels C, Anthuber M, Buettner R, and Jauch KW (2000). Overexpression of melanoma inhibitory activity (MIA) enhances extravasation and metastasis of A-mel 3 melanoma cells in vivo. Br J Cancer 83: 1216–1222.
Hemler ME, Huang C, Takada Y, Schwarz L, Strominger JL, and Clabby ML (1987). Characterization of the cell surface heterodimer VLA-4 and related peptides. J Biol Chem 262: 11478–11485.
Humphries MJ, Akiyama SK, Komoriya A, Olden K, and Yamada KM (1986). Identification of an alternatively spliced site in human plasma fibronectin that mediates cell type-specific adhesion. J Cell Biol 103: 2637–2647.
Jacob K, Bosserhoff AK, Wach F, Knuchel R, Klein EC, Hein R, and Buettner R (1995). Characterization of selected strongly and weakly invasive sublines of a primary human melanoma cell line and isolation of subtractive cDNA clones. Int J Cancer 60: 668–675.
Jaenicke R and Rudolph R (1986). Refolding and association of oligomeric proteins. Methods Enzymol 131: 218–250.
Langholz O, Rockel D, Mauch C, Kozlowska E, Bank I, Krieg T, and Eckes B (1995). Collagen and collagenase gene expression in three-dimensional collagen lattices are differentially regulated by alpha 1 beta 1 and alpha 2 beta 1 integrins. J Cell Biol 131: 1903–1915.
Ledda F, Bravo AI, Adris S, Bover L, Mordoh J, and Podhajcer OL (1997). The expression of the secreted protein acidic and rich in cysteine (SPARC) is associated with the neoplastic progression of human melanoma. J Invest Dermatol 108: 210–214.
Loftus JC, Smith JW, and Ginsberg MH (1994). Integrin-mediated cell adhesion: The extracellular face. J Biol Chem 269: 25235–25238.
Lougheed JC, Holton JM, Alber T, Bazan JF, and Handel TM (2001). Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins. Proc Natl Acad Sci USA 98: 5515–5520.
Mohri H, Katoh K, Iwamatsu A, and Okubo T (1996). The novel recognition site in the C-terminal heparin-binding domain of fibronectin by integrin alpha 4 beta 1 receptor on HL-60 cells. Exp Cell Res 222: 326–332.
Mould AP, Komoriya A, Yamada KM, and Humphries MJ (1991). The CS5 peptide is a second site in the IIICS region of fibronectin recognized by the integrin alpha 4 beta 1. Inhibition of alpha 4 beta 1 function by RGD peptide homologues. J Biol Chem 266: 3579–3585.
Qian F, Vaux DL, and Weissman IL (1994). Expression of the integrin alpha 4 beta 1 on melanoma cells can inhibit the invasive stage of metastasis formation. Cell 77: 335–347.
Ruoslahti E (1996). RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol 12: 697–715.
Scott G, Ryan DH, and McCarthy JB (1992). Molecular mechanisms of human melanocyte attachment to fibronectin. J Invest Dermatol 99: 787–794.
Springer TA (1997). Folding of the N-terminal, ligand-binding region of integrin alpha-subunits into a beta-propeller domain. Proc Natl Acad Sci USA 94: 65–72.
Stoll R, Renner C, Zweckstetter M, Bruggert M, Ambrosius D, Palme S, Engh RA, Golob M, Breibach I, Buettner R, Voelter W, Holak TA, and Bosserhoff AK (2001). The extracellular human melanoma inhibitory activity (MIA) protein adopts an SH3 domain-like fold. EMBO J 20: 340–349.
Teixido J, Parker CM, Kassner PD, and Hemler ME (1992). Functional and structural analysis of VLA-4 integrin alpha 4 subunit cleavage. J Biol Chem 267: 1786–1791.
Terranova VP, Hujanen ES, Loeb DM, Martin GR, Thornburg L, and Glushko V (1986). Use of a reconstituted basement membrane to measure cell invasiveness and select for highly invasive tumor cells. Proc Natl Acad Sci USA 83: 465–469.
van Groningen JJ, Bloemers HP, and Swart GW (1995). Identification of melanoma inhibitory activity and other differentially expressed messenger RNAs in human melanoma cell lines with different metastatic capacity by messenger RNA differential display. Cancer Res 55: 6237–6243.
Varner JA, Emerson DA, and Juliano RL (1995). Integrin alpha 5 beta 1 expression negatively regulates cell growth: Reversal by attachment to fibronectin. Mol Biol Cell 6: 725–740.
Wach F, Eyrich AM, Wustrow T, Krieg T, and Hein R (1996). Comparison of migration and invasiveness of epithelial tumor and melanoma cells in vitro. J Dermatol Sci 12: 118–126.
Woods A, McCarthy JB, Furcht LT, and Couchman JR (1993). A synthetic peptide from the COOH-terminal heparin-binding domain of fibronectin promotes focal adhesion formation. Mol Biol Cell 4: 605–613.
Acknowledgements
We are indebted to Dr. Brigitte Kaluza and Martin Kaufmann (Roche Diagnostics) for providing purified monoclonal MIA antibodies, recombinant MIA protein, and the MIA ELISA; to Ute Straube, Astrid Hamm, Selma Huber, and Ines Breibach for technical assistance; to Dr. Eberhard Klein (University of Würzburg) for providing αvβ3 and α6 integrin antibodies; and to Dr. Meenhard Herlyn for critically reading the manuscript. RS gratefully recognizes support from the German Academic Exchange Service (DAAD) and the German Research Foundation (DFG).
This work was supported by grants from the German Research Foundation (DFG) and the German Cancer Aid (Deutsche Krebshilfe) to AB and RB, and by a grant from the German Research Foundation to TAH (SFB469).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bosserhoff, AK., Stoll, R., Sleeman, J. et al. Active Detachment Involves Inhibition of Cell-Matrix Contacts of Malignant Melanoma Cells by Secretion of Melanoma Inhibitory Activity. Lab Invest 83, 1583–1594 (2003). https://doi.org/10.1097/01.LAB.0000097191.12477.5D
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.LAB.0000097191.12477.5D
This article is cited by
-
Small Molecules Antagonise the MIA-Fibronectin Interaction in Malignant Melanoma
Scientific Reports (2016)
-
Update of molecular pathobiology in oral cancer: a review
International Journal of Clinical Oncology (2014)
-
Protumoral roles of melanoma inhibitory activity 2 in oral squamous cell carcinoma
British Journal of Cancer (2013)
-
Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion
Nature Genetics (2013)
-
MIA is a potential biomarker for tumour load in neurofibromatosis type 1
BMC Medicine (2011)