Abstract
TRAIL (TNF-related apoptosis-inducing ligand) induces apoptosis by cross-linking of the two TRAIL receptors that contain a death domain, TRAIL-R1 and TRAIL-R2. TRAIL-R3 and TRAIL-R4 are receptors that do not transmit an apoptotic signal. Our aim was to determine the expression of TRAIL and its receptors in normal pancreas and chronic pancreatitis. We applied real-time PCR, immunohisto(cyto)chemistry, and nick-end labeling of apoptoses. In normal pancreas, a minor subset of acinar cells coexpressed TRAIL-R2 and TRAIL-R4, whereas ductular epithelium and interstitial fibroblast-like cells (FLC) expressed TRAIL-R4. TRAIL-R1 and TRAIL-R3 were not detected in normal pancreas. In chronic pancreatitis, the exocrine epithelium strongly expressed TRAIL-R1, -R2, -R4, and, to a lesser extent, TRAIL-R3. Islets focally neoexpressed TRAIL-R1 and -R2 and intensely expressed TRAIL-R4. Changes in TRAIL receptor expression were most pronounced in areas of inflammatory infiltration and active fibrosis. In normal pancreas, expression of TRAIL was low on the mRNA level and undetectable on the protein level. In chronic pancreatitis, FLC in areas of active fibrosis expressed TRAIL. In addition, apoptoses were most numerous in these areas. We show that these FLC are pancreatic stellate cells. Pancreatic stellate cells express TRAIL in vivo and in vitro, and TRAIL expression is enhanced by IFN-γ. Our findings indicate that the TRAIL/TRAIL receptor system is likely to be involved in chronic pancreatitis and suggest that pancreatic stellate cells may directly contribute to acinar regression by inducing apoptosis of parenchymal cells in a TRAIL-dependent manner.
Similar content being viewed by others
Introduction
TRAIL (Apo-2L) is an apoptosis-inducing ligand of the TNF superfamily (Marsters et al, 1997; Pitti et al, 1996; Walczak and Krammer, 2000; Wiley et al, 1995). Like other members of this family, TRAIL exists in two forms—a membrane-bound form and a soluble form—after proteolytic cleavage by cysteine proteases. TRAIL mRNA was detected in many normal human tissues in varying amounts, with very low levels in pancreas (Ozawa et al, 2001; Wiley et al, 1995). So far, four homologous TRAIL receptors have been characterized, two of which, TRAIL-receptor 1 (TRAIL-R1/DR4) and TRAIL-R2 (APO-2/DR5/KILLER/TRICK2), bear cytoplasmic death domains and mediate apoptosis (Pan et al, 1997b; Screaton et al, 1997; Sheridan et al, 1997; Walczak et al, 1997; Wu et al, 1997). Two other receptors, TRAIL-R3 (DcR1/TRID) and TRAIL-R4 (-R4/DcR2/TRUNDD), lack a functional death domain, and it has been proposed that they act as “decoy” receptors (Degli-Esposti et al, 1997a, 1997b; Marsters et al, 1997; Pan et al, 1998). By Northern blot analysis, it has been shown that on the mRNA level, TRAIL-R1 (Pan et al, 1997b), TRAIL-R2 (Pan et al, 1997a; Screaton et al, 1997; Sheridan et al, 1997; Walczak et al, 1997; Wu et al, 1997), and TRAIL-R4 (Marsters et al, 1997) are widely expressed in normal tissues. In contrast, TRAIL-R3 mRNA expression was reported to be much more restricted, because it was mainly detected in peripheral mononuclear cells and spleen (Degli-Esposti et al, 1997b). Osteoprotegerin is a soluble receptor for TRAIL, which has been shown to play a role in osteogenesis and inhibition of apoptosis in Jurkat cells (Emery et al, 1998). Normal human pancreas (NP) contained no or very low levels of TRAIL-R1 and -R3 transcripts (Ozawa et al, 2001) and low amounts of mRNA for TRAIL-R2 (Ozawa et al, 2001; Pan et al, 1997a; Sheridan et al, 1997), whereas TRAIL-R4 transcripts were detected at very low (Marsters et al, 1997) or intermediate levels (Pan et al, 1998). To date, no systematic and detailed analysis of expression of TRAIL and its receptors in NP has been performed and, hence, there is no knowledge of the role this death receptor/ligand system may play under normal and reactive, eg, inflammatory conditions.
We show here that NP is generally devoid of TRAIL and its receptors. Yet, under chronic inflammatory conditions as encountered in chronic pancreatitis (CP), TRAIL receptors are extensively and differentially induced by resident pancreatic cells, especially in areas of active inflammation and fibrosis. In CP, TRAIL is induced in scattered exocrine epithelia and pancreatic stellate cells (PSC). Furthermore, TRAIL expression in cultured human PSC is significantly increased by IFN-γ, a potential inflammatory cytokine that we have previously shown to be present at elevated tissue levels in CP (Hasel et al, 2001).
Results
General Aspects
Specimens from patients with CP were markedly microheterogeneous with respect to TRAIL receptor expression. This heterogeneity was paralleled by the microheterogeneity of inflammatory activity and parenchymal remodeling caused by the underlying disease. In other words, in microareas only mildly affected by CP, the TRAIL receptor profile very much resembled that of NP, whereas TRAIL receptor induction was at its maximum in areas of dense lymphohistiocytic infiltration and/or early fibrosis. Table 1 lists the maximum induction in a given cell type.
TRAIL-R1
In NP, TRAIL-R1 mRNA was detectable at very low levels, whereas in CP TRAIL-R1 transcript levels were found to be slightly higher (Fig. 1). In NP, TRAIL-R1 protein was undetectable by immunohistology (Table 2, Fig. 2). In CP, there was a considerable induction of TRAIL-R1 antigen in the epithelial compartment of the glands. Cells of ductules and tubular acini were strongly TRAIL-R1 positive; normal acini and duct epithelium were positive throughout, although less intensely. Islets were mostly negative or weakly TRAIL-R1 positive in advanced fibrotic areas (Fig. 2).
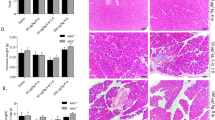
Semiquantitative RNA analysis. Cyclophilin was used as the internal standard. The diagram depicts the ×-fold expression of TRAIL, TRAIL-R1, -R2, -R3, and -R4 in chronic pancreatitis (CP) compared with normal pancreatic tissue, set as 1, after normalization of the CT values of each sample to its internal standard and calibration of CP to normal pancreas (NP).
Micrographs on the left show NP and, on the right, tissue affected with CP. TRAIL-R1: NP is immunohistologically devoid of TRAIL-R1 (original magnification, × 57). In CP, TRAIL-R1 is expressed in all pancreatic epithelia, although at varying antigenic densities. Note that islet cells are stained weakly compared with exocrine epithelia (original magnification, × 57). TRAIL-R2: In NP, TRAIL-R2 is weakly expressed in a subset of acinar and ductular cells, whereas all other cell types are TRAIL-R2 negative (original magnification, × 57). In CP, TRAIL-R2 is expressed in exocrine parenchyma, whereas islets are TRAIL-R2 negative in most instances, as depicted here (original magnification, × 114; inset, × 320). As a peculiar feature of columnar epithelium of large ducts, TRAIL-R2 is localized at the basal and at the luminal cell surfaces. Furthermore, TRAIL-R2 is present in endothelial cells and in a subset of interstitial fibroblast-like cells (FLC) (original magnification, × 114). TRAIL-R3: NP is immunohistologically devoid of TRAIL-R3. The stained cells are intravascular granulocytes. This case had a conspicuous intravascular leucocytosis (original magnification, × 114). In CP, TRAIL-R3 is inconsistently expressed in tubular acini epithelium. In cases expressing TRAIL-R3, however, antigen density is sometimes high, as in this micrograph (original magnification, × 114). TRAIL-R4: In NP, overall TRAIL-R4 expression is generally low. The depicted case features TRAIL-R4 in just a few scattered interstitial FLC and intravascular granulocytes. In CP, TRAIL-R4 is heavily neoexpressed, the antigenic density being highest in islet cells and interstitial cells. Note a gradient in TRAIL-R4 expression of acini ranging from strongly positive in the close vicinity of inflammatory infiltrates and areas rich in FLC, to negative in unaffected parenchymal microareas. In parenchymal areas with lymphohistiocytic infiltration and active fibrosis, nearly every cell expresses TRAIL-R4 at high levels (original magnification, × 57). TRAIL: NP is immunohistologically devoid of TRAIL (original magnification, × 57). In CP, TRAIL is inconsistently expressed in peri-insular FLC of islands in fibrotic areas (original magnification, × 114). In areas with heavy inflammatory infiltration, TRAIL is inconsistently detectable in tubular acini and ductular epithelia (original magnification, × 114). TRAIL is often detectable in interstitial FLC of areas of active fibrosis. Note a very weak TRAIL positivity in ductular structures (original magnification, × 114).
TRAIL-R2
TRAIL-R2 transcripts were present in both NP and CP. However, TRAIL-R2 mRNA levels were consistently higher in CP (Fig. 1). At the protein level, a smaller subset of acinar and ductular cells were TRAIL-R2 positive (Table 2), with no obvious association with structural features of NP (Fig. 2). In CP, the exocrine epithelium was entirely TRAIL-R2 positive; islets were mostly negative (Fig. 2) or weakly positive. Furthermore, TRAIL-R2 was present in endothelial cells, nerves, and a subset of interstitial fibroblast-like cells (FLC) and lymphocytes. In duct cells, TRAIL-R2 was located at the basal and luminal cell membranes (Fig. 2).
TRAIL-R3
TRAIL-R3 transcripts were undetectable in NP but were found in CP specimens (Fig. 1). At the antigen level, NP were TRAIL-R3 negative, except for intravascular granulocytes, which were strongly positive. Yet, for unknown reasons—most likely because of surgical trauma—one of the NP specimens contained foci of numerous intravascular granulocytes (Fig. 2). In CP, there was only a focal induction of TRAIL-R3 in resident cells. A subset of tubular acini (Table 2, Fig. 2) showed varying intensities of TRAIL-R3 positivity. TRAIL-R3 was further detected in subsets of acini and interstitial FLC.
TRAIL-R4
TRAIL-R4 was expressed on the mRNA level in each pancreas tested, although expression levels rose dramatically in CP (Fig. 1). At the protein level, TRAIL-R4 was expressed in a minor subset of acini and in interstitial FLC in NP. Furthermore, a subset of intravascular granulocytes expressed TRAIL-R4 protein (Table 2, Fig. 2). In CP, there was an extensive increase of TRAIL-R4 in numerous cell types. The inflammatory infiltrate, interstitial FLC, and endothelial cells strongly expressed TRAIL-R4. Also pancreatic islets were found to be TRAIL-R4 positive throughout, often very strongly so (Fig. 2), whereas in the exocrine compartment TRAIL-R4 expression was very inhomogeneous. Ductular cells were strongly positive throughout; duct and acinar cells ranged from TRAIL-R4 negative to strongly TRAIL-R4 positive. As a peculiar feature, there was a gradient of emergence of TRAIL-R4 in these cell types, with a maximum in close vicinity to inflammatory cells and in areas with active fibrosis, in contrast to a minimum in unaffected parenchymal areas (Fig. 2). Among CP specimens, expression of TRAIL-R4 by endocrine and exocrine epithelia correlated with the severity of inflammation and fibrosis. We observed cases showing an overall and strong expression of TRAIL-R4 in nearly every cell. But, even in these massively induced tissues, nerves and interstitial or peripancreatic adipocytes remained TRAIL-R4 negative.
TRAIL
Against the background of constitutive paucity of TRAIL receptors expressed in NP, CP was characterized by a differential and partially strong neoexpression of mainly TRAIL-R2 and -R4 and, less so, of TRAIL-R1 and -R3 by different types of resident and infiltrating cells. The question thus arose as to whether TRAIL is locally produced and, if so, by which type(s) of cell(s). Low levels of TRAIL transcripts were present in NP and slightly increased in CP (Fig. 1). Immunohistologically, TRAIL was undetectable in all specimens of NP (Table 2, Fig. 2). In CP, TRAIL protein was expressed at mostly low or very low (Fig. 2) levels in a minor subset of tubular acini and in ductular and duct epithelium. The cell type that exhibited the most distinctive TRAIL expression had a fibroblast-like appearance. These TRAIL-expressing FLC were rarely located around islets surrounded by enhanced connective tissue but were regularly found in areas rich in interstitial FLC and active fibrosis.
Apoptosis Preferentially Occurs in Areas of Active Fibrosis
On the one hand, the high expression levels of TRAIL receptors in pancreatic resident cells in areas of inflammatory infiltrates and in areas of active fibrosis and, on the other hand, TRAIL expression in epithelial cells of ductuli and tubular acini and interstitial FLC in CP, led us to study the localization of apoptotic cells in both NP and CP. By TUNEL (terminal deoxyribonucleotidyl transferase-mediated biotin-UTP nick-end labeling) staining we only rarely found apoptotic bodies in NP and, when present, these TUNEL-positive nuclei were located in vital, nonapoptotic acinar and duct cells suggesting phagocytosis of apoptotic bodies by these epithelia (data not shown). Yet in CP, TUNEL-positive nuclei were clearly more numerous and preferentially occurred at three sites: in areas rich in FLC (Fig. 3), less frequently within and beneath the epithelial layer of dilated ducts (not shown), and rarely in areas of inflammatory infiltration (not shown).
Apoptotic events in CP, as revealed by TdT-mediated biotin-UTP nick-end labeling. A, Overview of a parenchymal area characterized by active fibrosis and paucity of inflammatory cells showing numerous apoptoses (original magnification, × 57). B, Stained apoptotic bodies are located at the interphase of parenchymal lobuli and stroma rich in interstitial FLC (original magnification, × 114).
TRAIL Is Expressed in PSC In CP
To determine whether TRAIL is also expressed in PSC located in areas of active fibrosis, double immunofluorescence for TRAIL (FITC) and α-smooth muscle actin (tetramethylrhodamine isothiocyanate), a marker for PSC, was performed. This double labeling revealed a strong, almost exclusive staining for both antigens in grouped FLC of fibrotic areas surrounding exocrine epithelium in tissues affected by CP (Fig. 4). Staining of normal pancreatic tissue revealed only weak TRAIL positivity in a few scattered acinar cells. There was no staining for α-smooth muscle actin in interstitial FLC (not shown).
Immunofluorescent detection of TRAIL-expressing pancreatic stellate cells (PSC) in tissues affected with CP by double staining of α-smooth muscle actin (red) and TRAIL (green) (A; original magnification, × 78). Most cells staining for α- smooth muscle actin also stained strongly for TRAIL showing a yellow color. Epithelial cells remained negative (B; original magnification, × 78; C; original magnification, × 314).
PSC Express TRAIL
Immunohistology revealed TRAIL expression in interstitial FLC. With respect to the tissue distribution of these cells in areas of active fibrosis, one possible candidate seemed to be the PSC. PSC are located in interlobular spaces and in interacinar regions (Bachem et al, 1998). We used cultured PSC and pancreatic fibroblasts grown on chamber slides. Using a TRAIL-specific mAb, we obtained positive stainings of variable intensity in a subset of cultured PSC (Fig. 5A) but not in fibroblasts (not shown). These data were confirmed by performing quantitative real-time PCR on four different PSC cultures for TRAIL mRNA expression compared with human embryonic kidney cells serving as a negative control and human embryonic kidney cells transfected with TRAIL encoding pcDNA3.1 as the positive control (Fig. 5B).
A, Immunocytologic detection of TRAIL expression in cultured PSC. A subset of cells is stained at variable intensities. B, Semiquantitative real-time PCR showed a 1.4-fold higher expression of TRAIL mRNA in PSC compared with pancreatic fibroblasts. Human embryonic kidney (HEK) cells transfected with full-length TRAIL were used as positive control; untreated HEK served as negative control.
TRAIL Expression Is Up-Regulated by IFN-γ
We previously showed that the local content of IFN-γ is significantly increased in CP (Hasel et al, 2001). We therefore stimulated human pancreatic stellate cells (hPSC) for 48 hours with IFN-γ and analyzed TRAIL mRNA expression by real-time PCR. Exposure of PSC to IFN-γ lead to a 9.2-fold increase (sem ± 0.26) in TRAIL mRNA expression compared with untreated controls. In contrast, expression of TRAIL receptors remained almost unaffected (TRAIL-R1 and -R2) or decreased slightly (TRAIL-R3 and -R4) (Fig. 6).
TRAIL protein content in cell lysates as well as in supernatants was significantly increased after 24 and 48 hours of IFN-γ treatment: specifically, in lysates, TRAIL protein content was 123.65 pg/ml (sem ± 0.46) at 24 hours and 145.45 pg/ml (sem ± 0.25) at 48 hours, whereas no TRAIL protein was detectable in untreated controls. TRAIL was also released into the supernatant, 113.10 pg/ml (sem ± 0.20) at 24 hours and 119.8 pg/ml (sem ± 0.26) at 48 hours, whereas TRAIL was undetectable in supernatants of untreated controls. TRAIL expression and secretion did not affect the viability of PSC (not shown). Furthermore, TRAIL did not induce nuclear factor-κB (NF-κB) in these cells as evidenced by immunoblotting (Fig. 7). This suggests that there is no autocrine loop for TRAIL in PSC and that PSC-derived TRAIL has other targets.
NF-κB p65 protein expression in PSC controls and after 24 and 48 hours of treatment with IFN-γ. Jurkat cell lysate was used as a positive control. β-actin served as the internal loading control. Note the slight band within the nuclear protein preparation in PSC controls indicating a contamination with cytosolic proteins. ctr = control.
Discussion
NP contains transcripts for TRAIL receptors TRAIL-R1, albeit at very low levels (Ozawa et al, 2001), low amounts of mRNA for TRAIL-R2 (Ozawa et al, 2001; Pan et al, 1997a; Sheridan et al, 1997), and no detectable TRAIL-R3 transcripts (Sheridan et al, 1997), whereas TRAIL-R4 mRNA was detected at very low (Marsters et al, 1997) and intermediate levels (Pan et al, 1998). We show that this transcription profile corresponds to immunohistochemically detectable protein expression for TRAIL-R2, TRAIL-R3, and TRAIL-R4. Specifically in NP, TRAIL-R1 and TRAIL-R3 are not detectable in pancreatic resident cells; TRAIL-R2 is expressed in a minor subset of acinar and ductular epithelia, whereas TRAIL-R4 is expressed in interstitial FLC and in a minor subset of acinar cells. Thus, NP is largely devoid of TRAIL receptors.
We further show that this situation dramatically changes in the course of CP. In CP, there was a considerable induction of mainly TRAIL-R4 and TRAIL-R2, and less so, of TRAIL-R1 and TRAIL-R3 at both the mRNA and the protein level. In functional terms, exocrine and endocrine epithelia neoexpressed the apoptosis-mediating TRAIL-R1 and TRAIL-R2, together with the non–apoptosis-inducing receptors TRAIL-R4 and TRAIL-R3. Endothelial cells neoexpressed TRAIL-R2 and TRAIL-R4, vascular smooth muscle cells neoexpressed TRAIL-R4 in a subset of cells, nerves neoexpressed TRAIL-R2; interstitial FLC became partially TRAIL-R2 positive and TRAIL-R3 positive and featured enhanced TRAIL-R4 expression. This neoexpression of TRAIL receptors by pancreatic resident cells was observed in areas of lymphohistiocytic infiltration and also in areas of early, active fibrosis rich in FLC, ie, PSC. This suggests that the TRAIL-inducing stimuli may be delivered by the inflammatory infiltrate and/or PSC. On phenomenologic grounds, this was most obvious for TRAIL-R4 induction, because there was a gradient of parenchymal TRAIL-R4 positivity with its maximum adjacent to interstitial inflammatory cells and FLC and its minimum in healthy parenchymal areas. The sensitivity for the TRAIL-R4–inducing stimulus/stimuli may differ among the types of resident cells, because islets often expressed TRAIL-R4 at higher levels than surrounding acini and tubular acini, which stained weaker, even in areas of strong TRAIL-R4 positivity of neighboring cells. The histiocytic cells were devoid of TRAIL-R1, TRAIL-R2, and TRAIL-R3 in the presence of TRAIL-R4. Lymphocytes infiltrating the gland were negative for TRAIL-R1 and TRAIL-R3, and in part positive for TRAIL-R2 and TRAIL-R4. In accordance with published data (Degli-Esposti et al, 1997b; Pan et al, 1998), granulocytes strongly expressed TRAIL-R3 and were in part TRAIL-R4 positive. Adipocytes, being devoid of TRAIL receptors in NP, stayed negative in CP. Overall, the profound phenotypic change of a broad variety of cells brought about by factors initiating and maintaining CP (Etemad and Whitcomb, 2001), suggests that the expression of TRAIL receptors is differentially regulated.
At present, little is known about the molecular mechanisms that result in the induction of TRAIL receptor expression. Phillips et al (1999) reported that IFN-γ enhanced mRNA expression of TRAIL-R1 but not of TRAIL-R2, -R3, and -R4 in placental Hofbauer cells. Overexpression of TRAIL-R4 is reported to lead to NF-κB activation, suggesting protection against TRAIL-induced apoptosis (Degli-Esposti et al, 1997a). According to Hu et al (1999), neither overexpression of TRAIL-R4 nor NF-κB activation could prevent TRAIL-R1–induced apoptosis. IFN-γ has also been described to increase expression of TRAIL-R1 and TRAIL-R2 on the surface of human cytomegalovirus-infected human foreskin fibroblasts, whereas uninfected control cells showed a decrease in TRAIL-R1 and TRAIL-R2 surface expression upon IFN-γ stimulation (Sedger et al, 1999). IFN-γ and TNF-α enhanced TRAIL-R2 mRNA expression in a panel of cancers (Meng and el Deiry, 2001). In CP, tissue concentration of IFN-γ is increased compared with NP (Hasel et al, 2001). Its most likely source seems to be the lymphohistiocytic infiltrate, which mainly consists of CD3+CD4+ T cells and CD11c+, CD4+/−, S100p+ dendritic cells (Hasel et al, 2001). Thus, although IFN-γ may be a candidate cytokine for the local induction of TRAIL receptors in an inflammatory context, the question as to which might be the inducing factor(s) released by active fibrosis is still unanswered.
We also confirm and extend published data on expression of TRAIL in NP. In NP, TRAIL expression is detectable at the mRNA level (Pitti et al, 1996; Walczak and Krammer, 2000) but not by immunohistology. Although it is generally difficult to trace cytokines by immunohistology, we conclude that TRAIL is not a major constitutive product of normal pancreatic resident cells, unlike CD95L, another member of the TNF family of cytokines, which is constitutively expressed at high levels in normal pancreatic epithelia (Hasel et al, 2001). In CP, however, TRAIL was found in minor subsets of epithelial cells of tubular acini, ductules, and ducts. Immunohistochemistry failed to detect TRAIL in lymphocytes and histiocytic cells, although TRAIL has been found in these cells (Fanger et al, 1999; Kayagaki et al, 1999; Pitti et al, 1996; Santini et al, 2000). Again, levels of TRAIL expression by these cells might be below the threshold of immunohistology. However, TRAIL was clearly expressed in interstitial FLC in areas of active fibrosis. Thus, in tubular acini, ductules, and ducts, as well as in areas of active, early fibrosis, the neoexpression of TRAIL receptors by resident pancreatic cells locally coincides with the emergence of the ligand-producing cells. This local coincidence is mandatory for whatever consequence the engagement of TRAIL receptors by TRAIL may have in the context of CP. Interestingly, the frequency of apoptotic events in CP as revealed by TUNEL was highest in ectatic ducts and in areas with early fibrosis.
Thus, to better understand the pathophysiology of CP, it was important to exactly determine the nature of the interstitial FLC that express TRAIL in situ. We show here that at least one defined cell type that resides in areas of active fibrosis, namely the recently described PSC, a functional homolog to the hepatic “fat-storing” or Ito cell (Bachem et al, 1998; Schmid-Kotsas et al, 1999), expresses TRAIL in vivo and in vitro. As we further showed, these PSC represent the major subset of interstitial FLC in areas with active fibrosis. They are known to be fibrogenic in producing collagen types I and II, c-fibronectin, and laminin (Schmid-Kotsas et al, 1999). By local production of TRAIL, which is likely to be triggered by IFN-γ derived from lymphohistiocytic infiltrates (Hasel et al, 2001), PSC may in addition profoundly modify TRAIL-sensitive pancreatic resident cells, eventually enforcing apoptosis and, by this route, promoting parenchymal regression during CP.
Materials and Methods
Pancreatic Tissues
Twenty-six patients, whose informed consent was obtained before surgery, were chosen for this study. Twenty patients underwent partial pancreatectomy for CP: 12 were suffering from alcoholic pancreatitis, 6 had chronic nonalcoholic pancreatitis connected with pancreas divisum, 1 developed a cyst after initial surgery, and 1 had cryptogenic CP. Six patients underwent partial pancreatectomy for pancreas divisum. The specimens of these patients were histopathologically diagnosed as NP and included in the study as normal controls. All pancreatectomy specimens were immediately transferred to the laboratory where representative tissue samples were snap-frozen in liquid nitrogen and kept until further processing, ie, RNA and protein isolation using the methods described below. Other parts of the quick-frozen tissues were stored at −80° C, for later use in frozen section immunohistochemistry.
Isolation of hPSC
Human PSC cultures were isolated as described previously (Schneider et al, 2001). In short, surgically removed specimens from patients with CP were cut into small tissue blocks, cultured in 1:1 DMEM and HAM’s F12 supplemented with 2 mmol/L l-glutamine, penicillin/streptomycin, and amphotericin, and kept in an incubator at 37° C and 5% C02 humidified atmosphere. hPSC were separated by outgrowth for 1 to 3 days, seeded on glass coverslips, and grown up to 60% to 70% confluency.
mRNA Preparation and Semiquantitative Real-Time PCR
RNA from 6 tissue samples with NP and 18 with CP was prepared as described by Chomczynski and Sacchi (1987) and then digested with RNase-free DNase I (Boehringer Mannheim, Mannheim, Germany) and reverse transcribed using Superscript Reverse Transcriptase (Life Technologies, Gaithersburg, Maryland), random hexamer primer, and the addition of an RNase-inhibitor, following the manufacturer’s protocol. cDNA from PSC was produced as described previously (Schneider et al, 2001). Real-time semiquantitative analyses for TRAIL, TRAIL-R1, -R2, -R3, and -R4 mRNA were performed using the ABI PRISM 7700 Sequence Detection System instrument and software (PE Applied Biosystems, Foster City, California). Primers were designed by using Primer Express Software (PE Applied Biosystems) to meet the following criteria: not overlapping primers as close as possible to each other, avoidance of sequences with propensity for dimer formation, low G-C content, avoiding run of nucleotides, and a Tm at around 60° C. The sequences of the primer pairs are shown in Table 1. Primer pairs were purchased from MWG Biotech (Ebersberg, Germany). Cyclophilin expression was chosen as the endogenous control (forward primer: 5′-ATGGTCAACC CCACCGTGT-3′; reverse primer: 5′-TCTGCTGTCTTTGGGACCTTGTC-3′).
First the absence of nonspecific amplification was confirmed by analyzing the PCR amplification products by agarose gel electrophoresis. Amplicons generated from cDNA were also tested against no template control and RNA. In a further step, optimal primer concentrations were selected by mixing primer concentrations from 50 to 900 nmol of each forward and reverse primer. The curves were checked for low CT, for fast rising, and for confirmation analyzed by agarose gel electrophoresis. Real-time PCR was performed using 4 μl of cDNA (12.5 ng/μl), 4 μl of primer mix (forward/reverse) at optimal concentrations, 12 μl of sterile distilled water, and 20 μl of SYBR Green PCR Master Mix (PE Applied Biosystems) per reaction. The following cycling conditions were set: denaturation at 95° C for 2 minutes, followed by 40 cycles at 95° C for 15 seconds and 60° C for 1 minute.
Gene expression of TRAIL and TRAIL receptors of tissue affected with CP was measured relative to NP as the calibrator sample. All quantitations were also normalized to cyclophilin as an endogenous control to account for variability in the initial concentration of total RNA.
Analysis of quantitation was performed by calculating the following: (1) mean CT value of two replicates per sample; (2) difference between mean CT values of samples for each target (CP) and those of the endogenous controls (CT); (3) difference between mean CT values of the samples for each target (CP) and the mean CT value of the corresponding calibrator (NP) (CT). The quantitation is expressed as 2−CT, to allow graphical presentation, and shown as ×-fold expression of the target gene in CP compared with NP set as 1.
Immunohistochemistry
Immunohistochemistry was performed on tissues of 4 NP and 14 pancreases affected by CP. Serial, 2-μm-thick cryosections were immediately fixed in ice-cold acetone for 10 minutes, air dried, and incubated for 1 hour with mouse anti-human mAb in appropriate dilutions. Anti-TRAIL-R1 (clone M272, IgG1 isotype), anti-TRAIL-R2 (clone M412, IgG1 isotype), anti-TRAIL-R3 (clone M430, IgG1 isotype), anti-TRAIL-R4 (clone M440, IgG2a isotype), and anti-TRAIL (clone M182, IgG1 isotype) were provided by Immunex (Seattle, Washington). These mAb have been shown to be antigen specific and have previously been used for Western blotting, functional studies (Griffith et al, 1999a, 1999b; Sedger et al, 1999; Sprick et al, 2000), flow cytometry (Sprick et al, 2000), and immunohistology (Sträter et al, 2002). Bound primary antibody was detected via goat anti-mouse immunoglobulins conjugated to peroxidase labeled-dextran polymer in Tris-HCL buffer containing carrier protein (EnVision, Dako). 3-amino-9-ethyl-carbazole (Sigma, St. Louis, Missouri) was used as the substrate. Counterstaining was done with hemalaun. Cultured PSC grown on glass coverslips were washed twice in PBS, fixed for 30 minutes in −20° C acetone, and then immunostained with the anti-TRAIL mAb as described above. Controls were performed by omitting the first step antibody and yielded negative results. To determine the antigen specificity of the anti-TRAIL mAb used, TRAIL-transfected cells grown on cover slides were incubated with the primary antibody as described above. Cells transfected with TRAIL-R3 served as a negative control. Binding of primary antibody was visualized using biotinylated anti-mouse immunoglobulins from sheep and a streptavidin peroxidase system (Amersham, Buckinghamshire, United Kingdom). 3-amino-9-ethyl-carbazole was used as the substrate. Counterstaining was done with hemalaun (not shown).
Double immunofluorescence for detection of TRAIL expressing PSC was performed using mouse mAb against human α-smooth muscle actin (clone 1H5, IgG2a; Dako) as a marker for PSC, followed by a tetramethylrhodamine isothiocyanate-labeled Ig2a-specific goat anti-mouse secondary antibody and repeated washing. TRAIL was detected as described above; as the secondary antibody, an Ig1-specific FITC-labeled goat anti-mouse mAb was applied. Nuclear staining was done using 4,6-diamidino-2-phenylindole.
Human Soluble TRAIL ELISA
To determine protein concentrations of TRAIL in PSC, cells were treated with IFN-γ for 24 and 48 hours. Additionally, supernatants were sampled. We applied a commercial ELISA kit including a standard of known TRAIL concentration (EuroClone, Devon, United Kingdom). A total of 100 μl of each protein lysate together with a dilution series of the standard were dispensed in triplicate in 96-well microtiter plates precoated with an mAb against human TRAIL for antigen capture. Simultaneously, a biotinylated mAb specific against human TRAIL IFN-γ was added for detection. Plates were sealed and incubated for 3 hours at room temperature. After repeated washing, streptavidin-conjugated horseradish peroxidase was added and plates were resealed, incubated for 30 minutes at room temperature, and then washed again. A ready-to-use TMB solution was used as the substrate. This reaction leads to the formation of a colored product absorbing light at 450 nm. The color reagent was dispensed in each well. The light absorption was measured and calibrated against the standard using a MRX microplate reader (Dynatech Laboratories, Chantilly, Virginia). All wells were measured in triplicate. Results were analyzed using the Revelation software (Dynatech) and were given in picograms per milliliter as means with standard deviations. The Wilcoxon test was applied for statistical comparison of values of untreated PSC, PSC treated with IFN-γ, and supernatants of treated and untreated cells.
TUNEL
Tissue specimens from NP and CP were fixed in 6% formalin, routinely processed, and embedded in paraffin wax. Sections of 2 μm were adhered to glass slides by incubating at 37° C overnight. Deparaffinization and nick-end labeling were performed as described previously (Sträter et al, 1995). In short, after deactivating endogenous peroxidase, slides were treated for 1 minute with microwave irradiation, then incubated with proteinase K (Merck, Darmstadt, Germany) to digest nuclear proteins. Terminal deoxyribonucleotidyl transferase (TdT) (Promega, Madison, Wisconsin) at a final concentration of 0.1 U/μl was used to insert Biotin-16-dUTP (Boehringer Mannheim) to the ends of the DNA fragments. Slides were developed using the streptavidin-peroxidase complex and 3-amino-9-ethylcarbazol as described above. Counterstaining was done with hemalaun. Negative controls were performed by omitting TdT.
Immunoblotting of NF-κB p65
PSCs were harvested after treatment with IFN-γ for 24 and 48 hours using a cell scraper. Cytosolic and nuclear protein preparations were performed as described by Dignam et al (1983). Protein concentration was determined using Bradford reagent (Bio-Rad, Munich, Germany). Fifty micrograms of cytosolic and nuclear protein was separated on a 10% to 20% Tricine precast gel (Novex, San Diego, California) and transferred to a polyvinylidene difluoride membrane. Membranes were incubated with anti-human NF-κB p65, IgG1 (PharMingen, San Diego, California) overnight after blocking in PBS containing 5% nonfat dried milk at 4° C. After washing membranes three times with PBS/0.02% Tween 20, they were incubated with sheep anti-mouse biotinylated immunoglobulins (1:5000) and streptavidin (1:5000) (Amersham, Arlington Heights, Illinois). Protein lysates of Jurkat cells served as positive control. After another three washes, the blots were developed by enhanced chemiluminescence using the ECL system (Amersham). To assure equal protein loading, membranes were washed again and reblotted with anti-human β-actin, IgG1 (Sigma) as described above.
References
Bachem MG, Schneider E, Gross H, Weidenbach H, Schmid RM, Menke A, Siech M, Beger H, Grunert A, and Adler G (1998). Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 115: 421–432.
Chomczynski P and Sacchi N (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159.
Degli-Esposti MA, Dougall WC, Smolak PJ, Waugh JY, Smith CA, and Goodwin RG (1997a). The novel receptor TRAIL-R4 induces NF-kappaB and protects against TRAIL-mediated apoptosis, yet retains an incomplete death domain. Immunity 7: 813–820.
Degli-Esposti MA, Smolak PJ, Walczak H, Waugh J, Huang CP, DuBose RF, Goodwin RG, and Smith CA (1997b). Cloning and characterization of TRAIL-R3, a novel member of the emerging TRAIL receptor family. J Exp Med 186: 1165–1170.
Dignam JD, Lebovitz RM, and Roeder RG (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489.
Emery JG, McDonnell P, Burke MB, Deen KC, Lyn S, Silverman C, Dul E, Appelbaum ER, Eichman C, DiPrinzio R, Dodds RA, James IE, Rosenberg M, Lee JC, and Young PR (1998). Osteoprotegerin is a receptor for the cytotoxic ligand TRAIL. J Biol Chem 273: 14363–14367.
Etemad B and Whitcomb DC (2001). Chronic pancreatitis: Diagnosis, classification, and new genetic developments. Gastroenterology 120: 682–707.
Fanger NA, Maliszewski CR, Schooley K, and Griffith TS (1999). Human dendritic cells mediate cellular apoptosis via tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J Exp Med 190: 1155–1164.
Griffith TS, Rauch CT, Smolak PJ, Waugh JY, Boiani N, Lynch DH, Smith CA, Goodwin RG, and Kubin MZ (1999a). Functional analysis of TRAIL receptors using monoclonal antibodies. J Immunol 162: 2597–2605.
Griffith TS, Wiley SR, Kubin MZ, Sedger LM, Maliszewski CR, and Fanger NA (1999b). Monocyte-mediated tumoricidal activity via the tumor necrosis factor-related cytokine, TRAIL. J Exp Med 189: 1343–1354.
Hasel C, Rau B, Perner S, Sträter J, and Möller P (2001). Differential and mutually exclusive expression of CD95 and CD95 ligand in epithelia of normal pancreas and chronic pancreatitis. Lab Invest 81: 317–326.
Hu WH, Johson H, and Shu HB (1999). Tumor necrosis factor-related apoptosis-inducing ligand receptors signal NF-κB and JNK activation and apoptosis through distinct pathways. J Biol Chem 274: 30603–30610.
Kayagaki N, Yamaguchi N, Nakayama M, Eto H, Okumura K, and Yagita H (1999). Type I interferons (IFNs) regulate tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) expression on human T cells: A novel mechanism for the antitumor effects of type I IFNs. J Exp Med 189: 1451–1460.
Marsters SA, Sheridan JP, Pitti RM, Huang A, Skubatch M, Baldwin D, Yuan J, Gurney A, Goddard AD, Godowski P, and Ashkenazi A (1997). A novel receptor for Apo2L/TRAIL contains a truncated death domain. Curr Biol 7: 1003–1006.
Meng RD and el Deiry WS (2001). p53-independent upregulation of KILLER/DR5 TRAIL receptor expression by glucocorticoids and interferon-gamma. Exp Cell Res 262: 154–169.
Ozawa F, Friess H, Kleeff J, Xu ZW, Zimmermann A, Sheikh MS, and Buchler MW (2001). Effects and expression of TRAIL and its apoptosis-promoting receptors in human pancreatic cancer. Cancer Lett 163: 71–81.
Pan G, Ni J, Wei YF, Yu G, Gentz R, and Dixit VM (1997a). An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277: 815–818.
Pan G, Ni J, Yu G, Wei YF, and Dixit VM (1998). TRUNDD, a new member of the TRAIL receptor family that antagonizes TRAIL signalling. FEBS Lett 424: 41–45.
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, and Dixit VM (1997b). The receptor for the cytotoxic ligand TRAIL. Science 276: 111–113.
Phillips TA, Ni J, Pan G, Ruben SM, Wei YF, Pace JL, and Hunt JS (1999). TRAIL (Apo-2L) and TRAIL receptors in human placentas: Implications for immune privilege. J Immunol 162: 6053–6059.
Pitti RM, Marsters SA, Ruppert S, Donahue CJ, Moore A, and Ashkenazi A (1996). Induction of apoptosis by Apo-2 ligand, a new member of the tumor necrosis factor cytokine family. J Biol Chem 271: 12687–12690.
Santini SM, Lapenta C, Logozzi M, Parlato S, Spada M, Di Pucchio T, and Belardelli F (2000). Type I interferon as a powerful adjuvant for monocyte-derived dendritic cell development and activity in vitro and in Hu-PBL-SCID mice. J Exp Med 191: 1777–1788.
Schmid-Kotsas A, Gross HJ, Menke A, Weidenbach H, Adler G, Siech M, Beger H, Grunert A, and Bachem MG (1999). Lipopolysaccharide-activated macrophages stimulate the synthesis of collagen type I and C-fibronectin in cultured pancreatic stellate cells. Am J Pathol 155: 1749–1758.
Schneider E, Schmid-Kotsas A, Zhao J, Weidenbach H, Schmid RM, Menke A, Adler G, Waltenberger J, Grünert A, and Bachem MG (2001). Identification of mediators stimulating proliferation and matrix synthesis of rat pancreatic stellate cells. Am J Physiol Cell Physiol 281: C532–C543.
Screaton GR, Mongkolsapaya J, Xu XN, Cowper AE, McMichael AJ, and Bell JI (1997). TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr Biol 7: 693–696.
Sedger LM, Shows DM, Blanton RA, Peschon JJ, Goodwin RG, Cosman D, and Wiley SR (1999). IFN-gamma mediates a novel antiviral activity through dynamic modulation of TRAIL and TRAIL receptor expression. J Immunol 163: 920–926.
Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI, Goddard AD, Godowski P, and Ashkenazi A (1997). Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 277: 818–821.
Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J, Krammer PH, and Walczak H (2000). FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 12: 599–609.
Sträter J, Koretz K, Gunthert AR, and Möller P (1995). In situ detection of enterocytic apoptosis in normal colonic mucosa and in familial adenomatous polyposis. Gut 37: 819–825.
Sträter J, Walczak H, Pukrop T, von Müller L, Kornmann M, Mertens T, and Möller P (2002). TRAIL and its receptors in the colonic epithelium: A putative role in the defense of viral infections. Gastroenterology 122: 659–666.
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA, Goodwin RG, and Rauch CT (1997). TRAIL-R2: A novel apoptosis-mediating receptor for TRAIL. EMBO J 16: 5386–5397.
Walczak H and Krammer PH (2000). The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp Cell Res 256: 58–66.
Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, and Smith CA (1995). Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 3: 673–682.
Wu GS, Burns TF, McDonald ER, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, Hamilton SR, Spinner NB, Markowitz S, Wu G, and el Deiry WS (1997). KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 17: 141–143.
Acknowledgements
The authors thank Ms. Anke Gruber, Mr. Heiko Stahl, Ms. Roswitha Rittelmann, and Ms. Simone Westenfelder for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by grants SFB518/A13 (to PM) and SFB518/A6 (to BR) from the Deutsche Forschungsgemeinschaft. HW was supported by the BioFuture Program of the BMF.
Rights and permissions
About this article
Cite this article
Hasel, C., Dürr, S., Rau, B. et al. In Chronic Pancreatitis, Widespread Emergence of TRAIL Receptors in Epithelia Coincides with Neoexpression of TRAIL by Pancreatic Stellate Cells of Early Fibrotic Areas. Lab Invest 83, 825–836 (2003). https://doi.org/10.1097/01.LAB.0000073126.56932.46
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.LAB.0000073126.56932.46
This article is cited by
-
β-Cell Dysfunction in Chronic Pancreatitis
Digestive Diseases and Sciences (2012)
-
Mechanisms of parenchymal injury and signaling pathways in ectatic ducts of chronic pancreatitis: implications for pancreatic carcinogenesis
Laboratory Investigation (2009)
-
Parenchymal regression in chronic pancreatitis spares islets reprogrammed for the expression of NFκB and IAPs
Laboratory Investigation (2005)