Abstract
In vitro model systems for studying uterine leiomyomas are limited in that human-derived leiomyoma cells grow poorly in culture compared with normal myometrial cells and begin to senesce early, at approximately passage 10 in our studies. To our knowledge, a good in vitro human-derived cell culturing system for leiomyomas does not exist. In an attempt to fill this void, we have immortalized a uterine leiomyoma cell line by inducing telomerase activity, which allows cells to bypass their normal programmed senescence. Telomerase activity was induced by infecting the target (uterine leiomyoma and normal myometrial) cells with a retroviral vector containing hTERT, the gene for the catalytic subunit of telomerase. Subsequent analysis by RT-PCR and the telomeric repeat amplification protocol assay confirmed expression of the inserted gene and induction of telomerase activity in leiomyoma and myometrial cells. Analysis of cells for estrogen receptor-α and progesterone receptor proteins by Western blotting showed no change in expression of these proteins between the immortalized and parental leiomyoma and myometrial cells. Both immortalized and parental myometrial and leiomyoma cells expressed the smooth muscle–specific cytoskeletal protein α-actin and were negative for mutant p53 protein as evidenced by immunocytochemical staining. The immortalized leiomyoma and myometrial cells showed no anchorage-independent growth, with the exception of a small subpopulation of immortalized leiomyoma cells at a higher passage that did form two to three small colonies (per 50,000 cells) in soft agar. None of the immortalized cells were tumorigenic in nude mice. In conclusion, our data show the successful insertion of the hTERT gene into leiomyoma and myometrial cells and the immortalization of these cell lines without phenotypic alteration from the parental cell types (up to 200 population doublings). These cells should help to advance research in understanding the molecular pathways involved in the conversion of a normal myometrial cell to a leiomyoma cell and the mechanisms responsible for the growth of uterine leiomyomas. Answers to these questions will undoubtedly lead to the development of more effective treatment and intervention regimens for clinical cases of uterine leiomyoma.
Similar content being viewed by others
Introduction
Uterine leiomyomas (fibroids; myomas) are the most frequently occurring tumor of the female reproductive tract and clinically affect at least 25% of American women of reproductive age (Crum, 1999; Lumsden and Wallace, 1998) and may affect 77% or more overall (Cramer and Patel, 1990). Although these benign uterine smooth muscle tumors rarely if ever progress to malignancy, they are associated with a number of reproductive problems such as infertility, lost pregnancy, pelvic pain, and menorrhagia and are the primary cause of hysterectomies in women in the United States (Buttram and Reiter, 1981; Lepine et al, 1997; Lumsden and Wallace, 1998). In search of alternative treatments, research has focused mainly on the hormone-responsive characteristics of leiomyomas (Friedman et al, 1993; Regidor et al, 1995; Rein et al, 1995; Rein and Nowak, 1992; Tiltman, 1997). While this research has furthered the understanding of steroid-regulated tumor growth, the molecular pathogenesis of the etiology of leiomyomas remains largely unknown (Stewart and Nowak, 1996; Tiltman, 1997). The elucidation of molecular pathways involved in the conversion of a normal myometrial cell to a leiomyoma cell or the mechanism(s) responsible for the excessive growth of uterine myometrial cells will undoubtedly lead to the development of more effective treatment and even prevention of the disease process.
Much of the in vitro research of the biology of human uterine leiomyomas has resulted from the use of explants or primary monolayer cultures established from tissue samples taken after hysterectomy or myomectomy (Fayed et al, 1989; Rein and Nowak, 1992). However, in vitro research using human-derived uterine leiomyoma cells can be challenging because these cells tend to grow poorly and senesce quickly in culture compared with uterine myometrial cells (Cramer et al, 1985; Moss and Benditt, 1975).
To overcome the challenges of working with explants or primary monolayer cultures of human leiomyoma cells, immortalized human leiomyoma and myometrial cell lines were created by infecting leiomyoma and myometrial cells with a retroviral vector containing hTERT, the gene for the catalytic subunit of telomerase (Nakayama et al, 1998). Without telomerase, cells lose DNA from the telomeres of the chromosomes with each replication until the telomere shortens to a critical length and the cells senesce. Telomerase is a ribonucleoprotein reverse transcriptase that stabilizes the telomere and chromosome structure by adding telomeric repeats that are lost during replication (Meyerson et al, 1997; Nakamura et al, 1997; Nakayama et al, 1998. The telomerase enzyme consists of three major components, hTR (human telomerase RNA), hTLP1 (human telomerase–associated protein), and hTERT or hTRT (also hEST2; human telomerase reverse transcriptase) (Nakayama et al, 1998). Most normal and neoplastic cells express hTR and hTLP1 (Ramakrishnan et al, 1998). Malignant cells also express high levels of hTERT, which seems to be sufficient for the cells to regain telomerase activity (Meyerson, 2000; Ulaner et al, 2000). On the other hand, most normal cells and benign neoplastic cells, including leiomyoma, do not possess telomerase activity (Meyerson, 2000; Ulaner et al, 2000; Zheng et al, 1997), although some express the hTERT gene at varying levels (Ramakrishnan et al, 1998; Ulaner et al, 2000).
Other successful inductions of telomerase activity in human somatic cells via introduction of hTERT have not led to phenotypic changes typical of malignant cells (Jiang et al, 1999; Morales et al, 1999; Nakayama et al, 1998). In this article we describe the insertion of the hTERT gene into leiomyoma and myometrial cells and describe morphologic, cellular, and biochemical phenotypic characteristics of these cells after immortalization with the hTERT gene.
Results
Establishment of Cloned Cell Lines
Molecular characterizations.
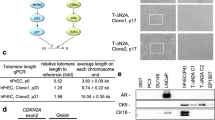
After G-418 antibiotic selection of successfully cloned cells, telomeric repeat amplification protocol (TRAP) analysis of the human uterine leiomyoma cell line GM10964 (UtLM), UtLM-hTERT, normal human uterine myometrial cells (UtSMC), and UtSMC-hTERT cells detected the presence of telomerase activity in the immortalized cells (hTERT) but not in the nonimmortalized cells (Fig. 1). Terminal restriction fragment (TRF) length analysis indicated that telomeres were lengthened in the immortalized cell lines after 50 population doublings, especially in the UtLM-hTERT cell line (Fig. 2). RT-PCR of the immortalized and nonimmortalized leiomyoma and myometrial cells for the hTERT gene showed expression of the gene only in the UtLM-hTERT and UtSMC-hTERT cell lines (Fig. 3). To ensure against false results from contamination by an outside cell type, the immortalized and nonimmortalized leiomyoma and myometrial cells were DNA fingerprinted. The UtLM and UtLM-hTERT showed the identical DNA fingerprint as did the UtSMC and UtSMC-hTERT (Fig. 4).
Telomerase activity of the human uterine leiomyoma cell line (UtLM), UtLM-human telomerase reverse transcriptase (hTERT), normal human uterine myometrial cells (UtSMC), and UtSMC-hTERT was determined by the telomeric repeat amplification protocol (TRAP) assay. Telomerase activity was present in UtLM-hTERT and UtSMC-hTERT. No telomerase activity was present in UtLM and UtSMC. Internal control consisted of a 10 attg internal telomerase assay standard (ITAS) DNA.
Terminal restriction fragment (TRF) length analysis of UtLM-hTERT and UtSMC-hTERT cells before infection (0) and at +2 and +50 population doublings after infection of the hTERT gene retrovirus. TRFs were qualitatively similar before and immediately after infection, but lengthened after 50 doublings, especially in the UtLM-hTERT cell line. PDLs = population doublings.
Phenotypic characterizations
Morphology and α-Smooth Muscle Actin Immunocytochemical Staining. The morphology of the UtLM-hTERT and UtSMC-hTERT remained the same as their nonimmortalized counterparts, UtLM and UtSMC, respectively (Fig. 5). Both the immortalized leiomyoma and myometrial cells were spindle with elongated, blunted nuclei typical of smooth muscle cells. These cells grew in a characteristic whorling pattern. Immunocytochemical staining revealed no difference in expression or localization of the cytoskeletal protein, α-smooth muscle actin, in respective nonimmortalized versus immortalized cells (Fig. 5).
Immunocytochemical staining of the cell lines UtLM (A), UtLM-hTERT (B), UtSMC (C) and UtSMC-hTERT (D) for α-actin cytoskeletal protein. Note cytoplasmic localization of α-actin in nonimmortalized (A and C) and immortalized (B and D) uterine leiomyoma (A and B) and smooth muscle (C and D) cells. 3,3′-diaminobenzidine tetrahydrochloride; ×150. Insets A, B, C, and D: Negative controls stained with nonimmune serum.
Estrogen Receptor-α (ER-α) and Progesterone Receptor (PR) Expression. Western blotting analysis for ER-α showed no difference in steroid hormone receptor expression between the immortalized and nonimmortalized cell lines. Signals at 67 kd for ER-α protein expression were slightly more intense on visual examination for the immortalized cell lines compared with the nonimmortalized cells (Fig. 6A). Three isoforms of the PR have been described, PR-A (120 kd), PR-B (94 kd), and PR-C (45–50 kd). All three isoforms were present in UtSMC and UtSMC-hTERT and in the positive control Ishikawa cell lines, but signals for PR-B were absent in UtLM and UtLM-hTERT cells. PR-A and PR-C were the predominant signals in all cell types compared with PR-B. The signal for PR-A was less in the UtLM and UtLM-hTERT cells than observed in immortalized or nonimmortalized UtSMC, although signals for PR-C were comparable (Fig. 6B).
Growth characteristics.
The nonimmortalized UtSMC grew at a similar rate to the UtSMC-hTERT, in which there was approximately 1 population doubling every 2 days; however, by Day 90, the UtSMC began to minimally “plateau,” whereas populations of UtSMC-hTERT actively doubled up to the last count of the study at Day 200 (Fig. 7). The nonimmortalized UtLM cells grew at a slower rate (approximately 1 population doubling every 10 days) than the immortalized UtLM-hTERT cells (1 population doubling every 3 days). By Day 30 the nonimmortalized UtLM cells began to plateau. Populations of the UtLM-hTERT actively doubled and had not reached a plateau by the last count at Day 200 (Fig. 7).
Malignant transformation characterizations.
Several indicators of malignancy were examined to observe whether induction of hTERT activity would result in tumorigenic transformation of the immortalized cell lines. UtLM, UtLM-hTERT, UtSMC, and UtSMC-hTERT were evaluated for their ability to grow in soft agar and nude mice and to express mutant p53 protein. Both immortalized and nonimmortalized leiomyoma and myometrial cells showed no anchorage-independent growth overall in soft agar after 3 weeks. However, a subpopulation of UtLM-hTERT cells (2 colonies/50,000 cells) at passage 46 formed colonies in soft agar (Fig. 8A). Subcutaneous injections of UtLM (data not shown), UtLM-hTERT (Fig. 8B), UtSMC (not shown), and UtSMC-hTERT (not shown) cell lines into intact female nude mice were negative for tumor formation. However, a malignant counterpart, the leiomyosarcoma cell line SK-LMS-1, produced large subcutaneous tumors (Fig. 8B). Both immortalized and nonimmortalized smooth muscle (not shown) and immortalized and nonimmortalized leiomyoma cell types were negative for mutated p53 expression compared with the positive control MCF7 cell line as evidenced by immunocytochemical staining (Fig. 9).
A, Soft agar assay. The cell lines UtSMC, UtSMC-hTERT, and UtLM did not form colonies in soft agar. The positive-control human leiomyosarcoma cells (SK-LMS-1) produced numerous colonies. The UtLM-hTERT cells (P 46) produced two small colonies (arrows). The negative controls (CCD-862SK human fibroblasts) showed no growth in soft agar. B, Nude mice injected subcutaneously in the right flank with 3 × 106 UtLM-hTERT or SK-LMS-1 (positive control) cells. At 8 weeks, tumor formation in the flank region of a mouse injected with SK-LMS-1 is shown.
Discussion
Expression of the catalytic subunit of telomerase, hTERT, and subsequent telomerase activity were successfully induced in a normal smooth muscle cell line and a benign smooth muscle neoplasm (leiomyoma) cell line. The mRNA for the hTERT gene and telomerase activity as measured by the TRAP assay are present in the transfected cell lines and not in the parental cell lines. TRF length analysis demonstrates that after several population doublings, the telomeres are lengthened in the immortalized cell lines and, in the UtLM-hTERT cells, the telomere length seems to become more uniform as indicated by the narrow band at +50 population doublings after infection (Fig. 2). These characteristics indicate that a stable transfection has occurred.
The aim of this study was to create a line of leiomyoma cells that can survive beyond normal programmed senescence and thus extend the usefulness of the cells for studying cellular mechanisms involved in the development and growth of uterine leiomyomas. This requires immortalization of the cell line without altering its phenotypic and genotypic characteristics. Several important phenotypic characteristics of the parental cell lines were unchanged in the cells expressing telomerase. The morphology of the immortalized cells remained the same as the nonimmortalized cells, and both the immortalized leiomyoma and myometrial cells showed typical spindled morphology and grew in a characteristic whorled pattern similar to what has been previously described for nonneoplastic smooth muscle and leiomyoma cells growing in culture (Moss and Benditt, 1975). Immunocytochemical staining revealed no difference in expression or localization of the smooth muscle–specific cytoskeletal protein, α-actin, in the UtSMC-hTERT and UtLM-hTERT versus the nonimmortalized parental cell lines. These results demonstrate that insertion of the hTERT gene into the UtLM and UtSMC cell lines and subsequent immortalization via induction of telomerase activity has not altered the morphologic characteristics of these cell lines.
Because leiomyomas are hormone-responsive tumors in vivo, and in particular are thought to be regulated in growth by estrogen and/or progesterone (Rein et al, 1995; Rein and Nowak, 1992; Tiltman, 1997), maintenance of ER and PR expression in the immortalized cell lines is important if these cells are to be used to study hormone responsiveness. We found that ER-α was present in all cell lines and that the pattern of expression of the various isoforms of PR did not differ between the immortalized cell lines and their respective parental lines.
In addition to evaluations for similarities to parental cell phenotype, the immortalized cells were also evaluated for transformation to an oncogenic phenotype. Restoration of telomerase activity is considered to be characteristic of spontaneously occurring malignancies. However, ectopic expression of telomerase through transfection of human telomerase catalytic component (hTERT) has been shown to drastically increase the number of possible population doublings without abnormally altering growth control or resulting in oncogenic transformation (Morales et al, 1999; Jiang et al, 1999). To date, however, these studies have involved normal (non-neoplastic) human cells. As with most normal human somatic cells, most benign neoplasms studied thus far, including uterine leiomyoma, have no telomerase activity. In fact the shortened telomeres found in leiomyomas when compared with adjacent normal myometrium in one study (Bonatz et al, 1998) demonstrates that these benign neoplasms are able to maintain their abnormal growth pattern without the aid of telomerase. Because an altered growth pattern already exists in these cells, however, transition to an oncogenic phenotype upon induction of telomerase activity could be easier in the leiomyoma cells than in normal somatic cells. Hahn et al (1999) have demonstrated that simultaneous expression of hTERT, oncogenic H-ras, and the simian virus 40 large-T oncoprotein in cultured human embryonic kidney cells and normal human fibroblasts results in malignant transformation of the cells, conferring to them the ability to form colonies in soft agar and tumors in nude mice.
With the insertion of the hTERT gene, the life span of the immortalized leiomyoma (UtLM-hTERT) and myometrial (UtSMC-hTERT) cells increases, yet the transfected cells do not seem to take on phenotypic characteristics of cancer cells. Both the immortalized myometrial and early passage immortalized leiomyoma cells do not show anchorage-independent growth and do not express mutant p53 protein, which are common characteristics of cancerous cells. Moreover, none of the immortalized cells produce tumors in nude mice, regardless of passage number. Results of the present study demonstrate that expression of the hTERT gene and subsequent induction of telomerase activity, by itself, in a benign uterine smooth muscle cell line (GM10964) does not induce oncogenic transformation, at least for lower passage number cells, and provides further evidence of the same effect in normal somatic cells (UtSMC).
The successful creation of immortalized leiomyoma and myometrial cell lines by insertion of the hTERT gene will be a huge advancement in the study of leiomyoma. The cells are a good model and show fidelity up to a certain number of passages. It seems that with time and increased replications, however, there may be changes in DNA stability or repair resulting in spontaneous changes/mutations. Further growth factor response and gene expression studies, as well as in depth genetic evaluations, will continue to validate the model and provide data toward the elucidation of molecular pathways involved in the conversion of a normal myometrial cell to a leiomyoma cell or the mechanisms responsible for the excessive growth of uterine smooth muscle cells. This will undoubtedly lead to the development of more effective treatment and intervention regimens for clinical cases of uterine leiomyoma. Additionally, the higher passage immortalized UtLM-hTERT cells may provide a model to help elucidate some of the molecular mechanisms of oncogenic transformation.
Materials and Methods
Cell Cultures
The UtLM cell line was obtained from Coriell Cell Repositories (Camden, New Jersey), and the cells were grown in minimum essential Eagle’s medium (M 5650; Sigma Chemical Company, St. Louis, Missouri) in 19% fetal bovine serum (FBS) (F-3885; Sigma) supplemented with vitamins (GIBCO BRL 11120-052; Grand Island, New York), essential and nonessential amino acids (GIBCO BRL 11130-051 and GIBCO BRL 11140-050, respectively), l-glutamine (GIBCO BRL 25030-081), and penicillin-streptomycin (GIBCO BRL 15070-063). Normal human uterine myometrial cells (UtSMC) were obtained from Clonetics Corporation (San Diego, California) and maintained in smooth muscle growth medium-2 (Clonetics CC-3181) supplemented with smooth muscle growth medium-2 singlequots (Clonetics CC-4149). Human ovarian adenocarcinoma cells (BG-1), breast cancer cells (MCF7), and endometrial cancer cells (Ishikawa) were grown in 1:1 DMEM-F12 supplemented with 10% FBS and 1% penicillin-streptomycin. Normal human skin fibroblasts (CCD-862SK) and human vulvar leiomyosarcoma cells (SK-LMS-1) were obtained from American Type Culture Collection (Rockville, Maryland; CRL-1784 and HTB-88, respectively) and maintained in the same media as the UtLM cells. The UtLM-hTERT cells were grown in the same media as UtLM cells with the exception of reduction of the FBS to 16% and replacement of penicillin-streptomycin with G-418 antibiotic (GIBCO BRL 11811–049) at 300 μg/ml. The UtSMC-hTERT cells were maintained in the same media as nontransfected uterine smooth muscle cells with the exception of the addition of G-418 (300 μg/ml) to the media. All cells were maintained and incubated for assays at 37° C and 5% CO2.
Insertion of hTERT into Cell Lines
The gene for hTERT was cloned into the pLXIN retroviral vector from Clontech (6062-1; Palo Alto, California) with the HpaI restriction enzyme. The cloned retrovirus was transfected into the RetroPack PT67 Packaging Cell Line (Clonetech K1060-D), where the vector was packaged into infectious, replication-incompetent retroviral particles. To infect the target cells, the packaged retrovirus (1 ml/dish) was then added to the medium of a 100-mm plate of target cells at 50% to 60% confluency. After 24 hours of incubation, the medium was changed, and the infected cells were selected by treatment with 300 μg/ml G-418 antibiotic. The pLXIN vector contains the neo gene, which confers resistance to the antibiotic G-418.
Telomerase Assay
Telomerase assays were performed with a PCR-based modified TRAP assay (Kim et al, 1994; Wright et al, 1995). The cell pellet (1 × 105) was suspended in 200 μl of cold TRAP lysis buffer, incubated for 30 minutes on ice, and centrifuged at 15,000 ×g for 20 minutes at 4° C. The extract equivalent of 103 cells (2 μl) was incubated with 48 μl of 20 mm Tris-HCl (pH 8.3), 1.5 mm MgCl2, 68 mm KCl, 0.05% Tween 20, 1 mm EGTA, 50 mm dNTP, 5 μg of BSA, 2 U of Taq DNA polymerase (Invitrogen, Carlsbad, California), 10 attg internal telomerase assay standard DNA, and 0.1 μg of extra purified TS primer (5′-AATCCGTCGAGCAGAGTT-3′) at 20° C for 30 minutes and then heated to 90° C for 3 minutes to terminate the telomerase reaction. During this step, 0.1 μg (2 μl) of extra purified CXII primer (5′-CCCTTACCCTTACCCTTACCCT-3′) was added, and the reaction mixture was subjected to 31 PCR cycles at 94° C for 30 seconds, 50° C for 30 seconds, and 72° C for 30 seconds (2 minutes for the final step). Ten microliters of PCR product was separated by electrophoresis on a 12% nondenaturing polyacrylamide gel and stained with SYBR Gold (Molecular Probes S11494; Eugene, Oregon) and detected by a fluorescence imaging system (Fuji FLA 3000; Fujifilm Medical Systems U.S.A., Inc., Stamford, Connecticut).
RT-PCR Analysis of hTERT
Total RNA was isolated using TRIZOL Reagent (GIBCO BRL 15596), a phenol-guanidine isothiocyanate solution, following the protocol supplied by GIBCO BRL. After isolation, the RNA was treated with DNase I (GIBCO BRL 18068-015) to remove any contaminating genomic DNA. We synthesized cDNAs from 1 μg of the isolated RNA with random primers and SuperScript II RNase H− Reverse Transcriptase (GIBCO BRL 18064-014). From the cDNAs, hTERT mRNA was amplified with the primer pair TRT/U1426 (5′-CCTCTGTGCTGGGCCTGGACGATA-3′) and TRT/L253 (5′-ACGGCTGGAGGTC TGTCAAGGTAG-3′), yielding a 282-bp amplification product. To control for intact cDNA, the mRNA for glyceraldehyde-3-phosphate dehydrogenase (G3PDH), a constitutively expressed enzyme, was amplified with the primer pair G3PDH-F (5′-ACCACAGTCCATGCCATCAC-3′) and G3PDH-R (5′-TCCACCACCCTGTTG CTGTA-3′). For all the amplifications, the reaction conditions were an initial denaturation at 94° C for 2 minutes followed by 30 cycles of denaturation at 94° C for 30 seconds and annealing/extension at 60° C for 1 minute, with a final extension step at 60° C for 7 minutes. The PCR products were fractionated on a 12% polyacrylamide gel, stained with SYBR Green I (Molecular Probes S7567), and analyzed with a UV transilluminating CCD imager.
TRF Length Analysis
TRF length analyses were performed as described previously (Tahara et al, 1997). Genomic DNA was purified by DNA extraction kits (Stratagene, LaJolla, CA). Three micrograms of genomic DNA was digested with the restriction endonuclease HinfI overnight at 37° C. The HinfI-digested DNA was resolved on 0.7% agarose gels and transferred to a nylon membrane (Hybond N, Amersham Corporation, Paisley, United Kingdom) under denaturing conditions. The membrane was hybridized with a 5′-32P–labeled (TTAGGG)4 telomeric oligonucleotide probe. Signal was detected by exposing the membrane to KODAK Bio Max film (Rochester, New York) with an intensifying screen.
DNA Fingerprinting
Cells were harvested from a 100-mm tissue culture dish, rinsed with PBS, resuspended in DNA lysis buffer (10 mm Tris-Cl, 1 mm EDTA [pH 8.0], 0.5% SDS and 100 μg/ml Proteinase K), and incubated at 55° C overnight. The lysate was then extracted with phenol:chloroform:iso-amyl alcohol (25:24:1); the top aqueous layer was recovered and DNA precipitated with 0.1 volume of 3 m sodium acetate (pH 5.2) and 2 volumes of ethanol. Precipitated DNA was redissolved in Tris-EDTA buffer (pH 8.0). The redissolved samples were then fingerprinted with the AmpliFLP D1S80 PCR Amplification Kit (Perkin-Elmer N808-0054; Norwalk, Connecticut) according to the manufacturer’s instructions.
Alpha-Smooth Muscle Actin (α-Actin) Expression by Immunocytochemical Staining
Uterine leiomyoma and normal myometrial cells (both immortalized and parent cell lines) were grown in chamber slides (Fisher Scientific 12-565-15; Pittsburgh, Pennsylvania) for in vitro staining and fixed with absolute methanol. Endogenous peroxidase activity was inactivated with 0.1% H2O2. Nonspecific staining was blocked by incubating the cells in normal goat serum (Vector Laboratories S-1000; Burlingame, California) diluted 1:60 in 0.01 m PBS. To detect the α-actin, we incubated the cells for 1 hour in supersensitive monoclonal mouse anti-α-actin primary antibody from BioGenex (AM128M-5; San Ramon, California) at a 1:10 dilution in 0.01 M PBS followed by a 30-minute incubation with a biotinylated goat anti-mouse secondary antibody (Supersensitive Multilink-HRP/DAB Kit from BioGenex, QD000-5L). After conjugation of streptavidin-horseradish peroxidase (BioGenex HK330-9K) to the secondary antibody, the complex was visualized with the chromogen, 3,3′-diaminobenzidine tetrahydrochloride (DAB) (Sigma D-9015). Cells were counterstained for 1 minute with diluted Mayer’s hematoxylin.
ER-α and PR Expression by Western Blotting Analysis
Total protein was collected from confluent T75 flasks of UtLM, UtLM-hTERT, UtSMC, UtSMC-hTERT, and Ishikawa cells for analysis of PR expression. After collection, the cells were washed three times with cold PBS-CMF (calcium-magnesium free PBS) and resuspended with 1 ml of cold isotonic lysis buffer (10 mm Tris [pH 8], 150 mm NaCl, 1% NP-40, 0.5% deoxycholate, 0.5% SDS, 50 μg/ml leupeptin, 100 μg/ml aprotinin, and 0.5 mm phenylmethylsulfonyl fluoride in double-distilled water). Keeping the samples on ice, the lysates were passed through 19-, 22-, and 25-gauge needles to mechanically break apart the cells. The lysates were then incubated on ice for 10 minutes and centrifuged at 14,000 rpm at 4° C for 20 minutes. The supernatant, containing the cellular protein, was collected, quantitated on a Beckman DU 650 Spectrophotometer (Palo Alto, California) using the Bio-Rad Protein Assay, and frozen at −80° C until time of immunoblotting. Nuclear protein was collected from confluent T75 flasks of UtLM, UtLM-hTERT, UtSMC, UtSMC-hTERT, and Ishikawa cells for analysis of ER-α expression. After collection, cells were washed three times with cold PBS-CMF and resuspended with 1 ml of cold low salt lysis buffer (20 mm Tris [pH 7.4], 5 mm KCl, 5 mm MgCl2, 0.5% NP-40, 0.1% deoxycholate, 0.1% SDS, 50 μg/ml leupeptin, 100 μg/ml aprotinin, and 0.5 mm phenylmethylsulfonyl fluoride in double-distilled water) to first extract the cytoplasmic protein. Keeping the samples on ice, the lysates were passed through 19- and 22-gauge needles to mechanically break apart the cytoplasm but not the nuclear envelope. The lysates were incubated on ice for 10 minutes and then centrifuged at 14,000 rpm at 4° C for 20 minutes. The supernatant, containing the cytoplasmic protein, was removed, leaving the nuclear pellet. To extract the nuclear protein, the nuclear pellet was resuspended in 500 μl of cold hypertonic lysis buffer (20 mm Tris [pH 8], 250 mm NaCl, 1% NP-40, 0.5% deoxycholate, 0.5% SDS, 50 μg/ml leupeptin, 100 μg/ml aprotinin, and 0.5 mm phenylmethylsulfonyl fluoride in double-distilled water). Keeping the samples on ice, the nuclear lysates were passed through 19-, 22-, and 25-gauge needles to mechanically break open the nuclear envelope. The lysates were incubated 10 minutes on ice and then centrifuged at 14,000 rpm at 4° C for 20 minutes. The supernatant, containing the nuclear protein, was collected and quantified as described above.
The Western blotting procedures used are described by Aoyama et al (1997). Equal amounts of protein were separated by SDS–PAGE on a 10% acrylamide gel, electroblotted onto a polyvinyldifluoride membrane (Immobilon-P, Millipore IPVH00010; Bedford, Massachusetts) and blocked overnight at 4° C. The membrane was incubated with 10 μg of the primary antibody, monoclonal mouse anti-human ER-α (ER1D5, Immunotech 1545, Rockford, Illinois) or PR (PR10A9, Immunotech 1546) for 2 hours at room temperature. Signal was detected with an ECL chemiluminescence kit (Amersham RPN2108) according to the manufacturer’s directions, using the anti-mouse secondary antibody at a 1:5000 dilution for 1 hour at room temperature.
Assay for Anchorage-Independent and Anchorage-Dependent Growth
Immortalized leiomyoma and myometrial cells, along with fibroblasts (CCD-862SK; negative controls) and SK-LMS-1 cells (positive controls) were seeded into 0.3% agar containing MEM and 10% FBS on top of a bed of 0.6% agar in 35-mm dishes at 5 × 104 cells per dish. Plating was done in duplicate. After 1 week of incubation (37° C and 5% CO2), approximately 0.5 ml of media was added to the dishes to avoid drying out and to ensure the cells had sufficient nutrients. Three weeks later, plates were stained overnight with a vital stain, 2-(4-isodophenyl)-3-(4-nitrophenyl)-5-phenyltetra-zolium chloride (Sigma 1-8377), and subsequently evaluated visually for the presence of colonies.
Nude Mouse Tumorigenicity Assay
The UtLM, UtSMC, UtLM-hTERT, and UtSMC-hTERT cells were suspended in 6 ml of PBS at a concentration of 1.5 × 107 cells/ml. Six-week-old female nude mice (Balb/c AnNCrl-nuBR) were given subcutaneous injections of 3 × 106 cells in the right flank. Nude mice injected in the flank with SK-LMS-1 (leiomyosarcoma) cells were used as positive controls. Negative controls consisted of nude mice injected in the flank region with human breast skin fibroblasts. The humane care and use of the mice were in accordance with institutional guidelines. The mice were observed daily. After 2 months, the mice were killed, and tumors were removed from mice injected with SK-LMS-1. The tumors were fixed in 10% neutral buffered formalin, paraffin embedded, sectioned at 6 μm, and stained with hematoxylin and eosin for histologic confirmation of tumor cell growth. A subset of mice injected with transfected and nontransfected leiomyoma and myometrial cells remained under study for a total of 4 months.
Mutant p53 Expression by Immunocytochemical Staining
Fixation and endogenous peroxidase inactivation, DAB chromogen visualization, and counter-staining procedures were similar to those described above for α-actin immunocytochemistry. Normal horse serum (1:60) (Vector Laboratories, Inc., Burlingame, California; S-2000) was used to block background staining. Cells were incubated with the primary antibody (p53 protein 1801, Novo Castra, NCL-p53–1801; Vector Laboratories, Inc.) at a 1:40 dilution for 2 hours, washed twice with PBS-CMF, and incubated with the secondary antibody (Vectastain Elite Mouse IgG, ABC kit; Vector Laboratories, Inc.) for 30 minutes.
References
Aoyama H, Kato H, and Dixon D (1997). Specificity of antibodies against rodent transforming growth factor-α protein. J Histochem Cytochem 45: 695–701.
Bonatz G, Frahm SO, Andreas S, Heidorn K, Jonat W, and Parwaresch R (1998). Telomere shortening in uterine leiomyomas. Am J Obstet Gynecol 179: 591–596.
Buttram VC Jr and Reiter RC (1981). Uterine leiomyomata: Etiology, symptomatology, and management. Fertil Steril 36: 433–445.
Cramer SF and Patel A (1990). The frequency of uterine leiomyomas. Am J Clin Pathol 94: 435–438.
Cramer SF, Robertson AL, Ziats NP, and Pearson OH (1985). Growth potential of human uterine leiomyomas: Some in vitro observations and their implications. Obstet Gynecol 66: 36–41.
Crum C (1999). The female genital tract. In: Cotran RS, Kumar V, and Collins T, editors. Pathologic basis of disease. Philadelphia: Saunders, 1035–1091.
Fayed YM, Tsibris JCM, Langenberg PW, and Robertson AL (1989). Human uterine leiomyoma cells: Binding and growth responses to epidermal growth factor, platelet-derived growth factor, and insulin. Lab Invest 60: 30–37.
Friedman AJ, Daly M, Juneau-Norcross M, Rein MS, Fine C, Gleason R, and Leboff M (1993). A prospective, randomized trial of gonadotropin-releasing hormone agonist plus estrogen-progestin or progestin “add-back” regimens for women with leiomyomata uteri. J Clin Endocrinol Metab 76: 1439–1445.
Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, and Weinberg RA (1999). Creation of human tumour cells with defined genetic elements. Nature 400: 464–468.
Jiang X-R, Jimenez G, Chang E, Frolkis M, Kusler B, Sage M, Beeche M, Bodnar AG, Wahl GM, Tlsty TD, and Chiu C-P (1999). Telomerase expression in human somatic cells does not induce changes associated with an immortalized phenotype. Nat Genet 21: 111–114.
Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, and Shay JW (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266: 2011–2015.
Lepine LA, Hillis SD, Marchbanks PA, Koonin LM, Morrow B, Kieke BA, and Wilcox LS (1997). Hysterectomy surveillance–United States. Mor Mortal Wkly Rep CDC Surveill Summ 46: 1–15.
Lumsden MA and Wallace EM (1998). Clinical presentation of uterine fibroids. Bailliéres Clin Obstet Gynaecol 12: 177–195.
Meyerson M (2000). Role of telomerase in normal and cancer cells. J Clin Oncol 18: 2626–2634.
Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Lui Q, Bacchetti S, Haber DA, and Weinberg RA (1997). hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90: 785–795.
Morales CP, Holt SE, Ouellette M, Kaur KJ, Yan Y, Wilson KS, White MA, Wright WE, and Shay JW (1999). Absence of cancer-associated changes in human fibroblasts immortalized with telomerase. Nat Genet 21: 115–118.
Moss NS and Benditt EP (1975). Human atherosclerotic plaque cells and leiomyoma cells. Am J Pathol 78: 175–185.
Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, and Cech TR (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277: 955–959.
Nakayama J-I, Tahara H, Tahara E, Saito M, Ito K, Nakamura H, Nakanishi T, Tahara E, Ide T, and Ishikawa F (1998). Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat Genet 18: 65–68.
Ramakrishnan S, Eppenberger U, Mueller H, Shinkai Y, and Narayanan R (1998). Expression profile of the putative catalytic subunit of the telomerase gene. Cancer Res 58: 622–625.
Regidor PA, Schmidt M, Callies R, Kato K, and Schindler AE (1995). Estrogen and progesterone receptor content of GnRH analogue pretreated and untreated uterine leiomyomata. Eur J Obstet Gynecol Reprod Biol 63: 69–73.
Rein MS, Barbieri RL, and Friedman AJ (1995). Progesterone: A critical role in the pathogenesis of uterine myomas. Am J Obstet Gynecol 172: 14–18.
Rein MS and Nowak RA (1992). Biology of uterine myomas and myometrium in vitro. Semin Reprod Endocrinol 10: 310–319.
Stewart EA and Nowak RA (1996). Leiomyoma-related bleeding: A classic hypothesis updated for the molecular era. Hum Reprod Update 2: 295–306.
Tahara H, Tokutake Y, Maeda S, Kataoka H, Watanabe T, Satoh M, Matsumoto T, Sugawara M, Ide T, Goto M, Furuichi Y, and Sugimoto M (1997). Abnormal telomere dynamics of B-lymphoblastoid cell strains from Werner’s syndrome patients transformed by Epstein-Barr virus. Oncogene 15: 1911–1920.
Tiltman AJ (1997). Smooth muscle neoplasms of the uterus. Curr Opin Obstet Gynecol 9: 48–51.
Ulaner GA, Hu J-F, Vu TH, Giudice LC, and Hoffman AR (1998). Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res 58: 4168–4172.
Ulaner GA, Hu J-F, Vu TH, Oruganti H, Giudice LC, and Hoffman AR (2000). Regulation of telomerase by alternate splicing of human telomerase reverse transcriptase (hTERT) in normal and neoplastic ovary, endometrium, and myometrium. Int J Cancer 85: 330–335.
Wright WE, Shay JW, and Piatyszek MA (1995). Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic Acids Res 23: 3794–3795.
Zheng PS, Iwasaka T, Yamasaki F, Ouchida M, Yokoyama M, Nakao Y, Fukuda K, Matsuyama T, and Sugimori H (1997). Telomerase activity in gynecologic tumors. Gynecol Oncol 64: 171–175.
Acknowledgements
We thank Drs. Gordon P. Flake and Cynthia A. Afshari for their critical review of this manuscript, and Mr. Norris D. Flagler for his expertise and assistance with the digital imaging.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Carney, S., Tahara, H., Swartz, C. et al. Immortalization of Human Uterine Leiomyoma and Myometrial Cell Lines After Induction of Telomerase Activity: Molecular and Phenotypic Characteristics. Lab Invest 82, 719–728 (2002). https://doi.org/10.1097/01.LAB.0000017499.51216.3E
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1097/01.LAB.0000017499.51216.3E
This article is cited by
-
The Effects of Periostin Expression on Fibroid-Like Transition of Myometrial Cells
Reproductive Sciences (2023)
-
Introduction of Somatic Mutation in MED12 Induces Wnt4/β-Catenin and Disrupts Autophagy in Human Uterine Myometrial Cell
Reproductive Sciences (2020)
-
H19 lncRNA identified as a master regulator of genes that drive uterine leiomyomas
Oncogene (2019)
-
Epigenetic regulation of transcription factor promoter regions by low-dose genistein through mitogen-activated protein kinase and mitogen-and-stress activated kinase 1 nongenomic signaling
Cell Communication and Signaling (2016)
-
Uterine fibroids are characterized by an impaired antioxidant cellular system: potential role of hypoxia in the pathophysiology of uterine fibroids
Journal of Assisted Reproduction and Genetics (2013)