Abstract
It has been previously shown that S100A2 is down-regulated in tumor cells and can be considered a tumor suppressor. We have recently shown that this down-regulation can be observed particularly in epithelial tissue, where S100A2 expression decreases remarkably in tumors as compared with normal specimens. In the present paper we investigate whether S100A2 could play a tumor-suppressor role in certain epithelial tissues by acting at the cell migration level. To this end, we made use of five in vitro human head and neck squamous cell carcinoma lines in which we characterized S100A2 expression at both RNA and protein level. To characterize the influence of S100A2 on cell kinetic and cell motility features, we used two complementary approaches involving specific antisense oligonucleotides and the addition of S100A2 to the culture media. The different expression analyses gave a coherent demonstration of the fact that the FADU and the RPMI-2650 cell lines exhibit high and low levels of S100A2 expression, respectively. Antisense oligonucleotides (in FADU) and extracellular treatments (in RPMI) showed that, for these two models, S100A2 had a clear inhibitory influence on cell motility while modifying the cell kinetic parameters only slightly. These effects seem to be related, at least in part, to a modification in the polymerization/depolymerization dynamics of the actin microfilamentary cytoskeleton. Furthermore, we found evidence of the presence of the receptor for advanced glycation end-products (RAGE) in RPMI cells, which may act as a receptor for extracellular S100A2. The present study therefore presents experimentally based evidence showing that S100A2 could play a tumor-suppressor role in certain epithelial tissues by restraining cell migration features, at least in the case of head and neck squamous cell carcinomas.
Similar content being viewed by others
Introduction
Calcium regulates a variety of biological processes as a second messenger, and Ca2+ signals are transduced into intracellular responses via an interaction with numerous Ca2+-binding proteins characterized by the EF-hand motif (Donato, 1999; Schäfer and Heizmann, 1996). The biological processes upon which calcium exerts a significant influence include cell proliferation, cell migration, and muscle contraction (Donato 1999; Gimona et al, 1997; Schäfer and Heizmann, 1996). The family of S100 proteins has grown to be one of the largest superfamilies of EF-hand proteins since the first S100 protein was identified from brain tissue more than 30 years ago as an acidic, low-molecular-mass, Ca2+-binding protein (Moore, 1965). The physiological and structural properties of S100 proteins suggest that they are trigger or activator proteins, whereas other Ca2+-binding proteins act mainly as buffers (Donato, 1999; Heizmann and Cox, 1998; Schäfer and Heizmann, 1996). To date, some 19 different proteins have been assigned to the S100 protein family, and the genes encoding more than ten S100 proteins are located in a cluster on human chromosome 1q21 (Schäfer and Heizmann, 1996).
S100A2 is of particular interest because its cDNA coding was identified in a search for novel tumor-suppressor genes as the result of subtractive hybridization between normal and tumor-derived human mammary epithelial cells (Lee et al, 1991). Extending this observation to breast tumors, Lee et al (1992) showed a marked down-regulation of S100A2 in breast tumor biopsies. In the same way, we have shown that, whereas no expression of the S100A2 gene could be detected on 11 tumorigenic human breast epithelial cell lines, a normal breast epithelial cell line and biopsy specimens did express S100A2 (Pedrocchi et al, 1994). The down-regulation of S100A2 is not restricted to breast cancer but is also found in melanomas (Maelandsmo et al, 1997) and other tumor types (Ilg et al, 1996). All these data have thus led to the designation of the S100A2 gene as a tumor suppressor, and S100A2 might therefore be generally down-regulated when cells acquire a tumorigenic phenotype, a factor which implies that the protein may play a role in inhibiting tumor progression or suppressing tumor cell growth (Wicki et al, 1997). In addition, the expression of S100A2 can be induced by growth factors (Lee et al, 1992; Stoll et al, 1998). For example, epidermal growth factor (EGF) receptor activation selectively stimulates S100A2 gene expression at the transcriptional level in human keratinocytes (Stoll et al, 1998).
In a recent study (N Nagy, D Hoyaux, I Gielen, BW Schafer, R Pochet, CW Heizmann, R Kiss, I Salmon, and C Decaestecker, unpublished data), we characterized the level of immunohistochemical S100A2 expression in a large set of normal and tumoral (benign and malignant) tissues of various histological origins, but epithelial in the great majority of cases (with either glandular, squamous, respiratory, or urothelial differentiation). The data show that S100A2 is expressed only marginally, if at all, in nonepithelial tissue. In epithelial tissue its level of expression decreases dramatically in tumors compared with normal specimens (Fig. 1A and B). We also showed that S100A2 is colocated with cytokeratin K14, an intermediate filament protein characteristic of epithelial cells, which is expressed in basal proliferative keratinocytes.
Panels A and B contain immunohistochemical illustrations of the level and pattern of expression of S100A2 in normal epithelial (A, × 200) and cancerous (B, × 400) tissue of the head and neck. Panels C to H contain immunofluorescence illustrations as follows: C to D (x1000), S100A2 expression (shown in red) in FADU cells cultured for 12 hours in the presence (0.1μm) of random control CL-A2 (C) or antisense AS-A2 (D) oligonucleotides (shown in green); E to F (× 400), glomerular actin cytoskeleton (red) and fibrillary actin cytoskeleton (green) in RPMI cells cultured for 24 hours in the absence (control, E) or the presence (F) of 1 nm S100A2; G to H (× 1000), binding of externally added S100A2 to RPMI cells: RPMI cells before (G) and 30 minutes after (H) the addition of S100A2 (10 nm) to the culture media. The S100A2 protein is revealed by red immunofluorescence, and DAPI staining (blue fluorescence) identifies the cell nuclei.
The present study aims to investigate whether S100A2 could play its tumor-suppressor role in epithelium tissue by acting at the cell-migration level. To this end, we first characterized the S100A2 expression levels in five in vitro human head and neck squamous cell carcinoma (HNSCC) lines (the FADU, Det-562, RPMI, SCC-9, and SCC-25 models). This was done at the RNA level by means of reverse transcription-polymerase chain reaction (RT-PCR) and at the protein level by means of Western blotting and immunohistochemistry. Two models (FADU and RPMI) expressing S100A2 at a high and a low level, respectively, were selected on the basis of the results obtained. The influence of S100A2 on these two models was then analyzed at the cell kinetic and cell motility levels. This was carried out in two complementary ways: S100A2 expression was inhibited in the FADU model (high natural expression) by means of antisense oligonucleotides, whereas S100A2 was added externally to the RPMI model (low natural expression). We also analyzed whether the S100A2 treatment was able to significantly modify the polymerization/depolymerization dynamics of the microfilamentary actin cytoskeleton, an important element in the dynamic changes involved in cell motility (Therior and Mitchison, 1991). Extracellular S100A2 effects were investigated in accordance with those already reported in the literature (Komada et al, 1996). We also investigated the presence of the receptor for advanced glycation end products (RAGE), a candidate receptor for S100/calgranulin polypeptides (Hofmann et al, 1999), which we found evidence of particularly in the RPMI model.
Results
S100A2 Expression in Five In Vitro Models of Human Squamous Cell Carcinomas
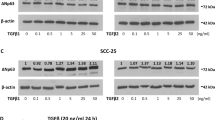
Taking the five HNSCC cell lines analyzed, Figure 2A and B, illustrates the results obtained by RT-PCR (A) to reveal the presence of S100A2 mRNAs and by Western blotting (B) to show S100A2 expression at the protein level. To confirm and refine these first results, quantitative analyses of S100A2 expression were undertaken by means of computer-assisted microscopy applied to conventional immunohistochemistry. In Figure 2C, the labeling index (LI) reports the percentage of immunopositive cells, and in Figure 2D, the mean optical density (MOD) denotes staining intensity computed on the immunopositive cells only. Because of standardization (see “Materials and Methods”), these two variables enable a precise quantitative evaluation and comparison of S100A2 expression to be established in the five cell lines analyzed. All of these data give a consistent representation of the different levels of S100A2 expression encountered in the five cell lines analyzed. Of these, the FADU and RPMI models exhibited particularly high and low levels of S100A2 expression, respectively. This is why we chose these two models to investigate the influence of S100A2 on HNSCC cell kinetic and migration properties, as reported below.
S100A2 expression in the five human head and neck squamous cell carcinoma (HNSCC) cell lines analyzed by means of different techniques. Results of the reverse transcription-polymerase chain reaction (RT-PCR) (A, including a glyceraldehyde-3-phosphate dehydrogenase [GADPH]-specific analysis as control as well as a negative control in column 6 of the gels), the Western blotting (B, where recombinant S100A2 at 1 ng/ml was used as positive control in column 6), and the quantitative immunochemical (C and D) analyses. Panel C reports the percentages of immunopositive cells (labeling index) and Panel D the staining concentrations (by means of the mean optical density variable) in the five cell lines quantified by means of computer-assisted microscopy.
S100A2 Inhibition in FADU Cells by Means of Specific Antisense Oligonucleotides
We chose the FADU and the RPMI cell lines as biological models. As reported above, while the RPMI model shows a relatively low level of expression, the FADU model shows a high one. We accordingly analyzed the effects of S100A2 inhibition in the FADU cells by means of antisense oligonucleotides and S100A2 added externally to the RPMI cells (see below).
Figure 1C and D, shows the decrease in S100A2 expression levels (red fluorescence) in the FADU cells caused by specific antisense oligonucleotides at 0.1 μm (Fig. 1C: CL-A2 condition; Fig. 1D: AS-A2 condition; oligonucleotides shown by green fluorescence). As shown in Figure 3A and B, the decrease in the level of S100A2 expression after oligonucleotide treatment (12 hours and 24 hours, respectively) was in fact specific at the smallest concentration (0.1 μm) only, ie, a very significant inhibition for AS-A2 and a lack of effect for CL-A2. At this small concentration, the antisense treatment appeared to be very efficient after 12 hours (Fig. 3A) and was maintained after 24 hours (Fig. 3B), but it was less efficient in terms of a decrease in the level of S100A2 expression. At 1 μm, a global “oligonucleotide” effect appeared, especially after 24 hours (Fig. 3B), but with a greater level of inhibition because of the antisense AS-A2. Similar observations were made for a concentration of 10 μm (data not shown). This seems to reflect the known side effects of oligonucleotide treatments. The antisense effect was confirmed on the neo-synthesis of the S100A2 protein by means of immunoprecipitation, as shown in Figure 3C. With respect to the control cells (ie, untreated cells), which were set to 100%, we observed a decrease of about 40% in the level of the immunoprecipitated S100A2 protein after 12 hours of treatment with 0.1 μm AS-A2 versus a lack of effect with CL-A2. A slight decrease (about 15%) was also observed with 0.01 μm AS-A2. Consequently, the 0.1 μm-oligonucleotide condition was used in the FADU cell motility assay reported below.
Effects of oligonucleotides (antisense S100A2: AS-A2 and random: CL-A2) on S100A2 expression in FADU cells. In Panels A and B, these effects (mean, sem and sd) have been evaluated by means of quantitative immunofluorescence (60 measures per condition, pooled from three independent experiments) after 12 hours and 24 hours of treatment respectively. Two different oligonucleotide concentrations of 0.1 and 1 μm are illustrated. Without any specific indication to the contrary, the p-values indicate the levels of significance (NS, not significant) in comparison with the control (CT). The p-values characterizing the differences between the AS-A2 and CL-A2 conditions at 1 μm are also indicated. Panel C confirms the oligonucleotide effects on the neosynthesis of the S100A2 protein by means of immunoprecipitation. FADU cells were metabolically labeled in the presence of 35S-cysteine and 35S-methionine during the final 5 hours of a 12-hour treatment with the oligonucleotides AS-A2 and CL-A2 (at 0.01 and 0.1 μm). Panel C shows the levels of immunoprecipitated S100A2 in the different conditions in relation to the levels in the untreated cells (control) set to 100%.
In Vitro Influence of S100A2 on the Cell Migration Features and Cell Kinetics of the FADU and RPMI Models
We used a phase-contrast, computer-assisted microscope to quantify both the cell kinetics and the cell migration features on living cells. This is illustrated in the eight digitized photographs shown in Figure 4, all of which were taken from an invariant point (not visible on these pictures but corresponding to the geometrical center of each one). This sequence begins at moment t = 30 hours, 00 minutes (ie, 30 hours after the beginning of the cell tracking procedure), and shows the evolutions of two cells labeled A and E over a period of 41 hours. Cell A gives birth to a generation of cells labeled A1 and A2 at time t = 30 hours, 35 minutes. Cell E gives birth to two generations of cells, E1 and E2, and then to E11 and E12, which are the daughter cells of E1. Cell E2 dies between time t = 35 hours, 10 minutes, and t = 43 hours, 00 minutes. Typical morphologies of dying cells are visible at time t = 33 hours, 30 minutes (Cell Dying 1, CD1), and their “ghosts” remain visible until time t = 44 hours, 10 minutes. Typical morphologies of mitoses are also illustrated in Figure 4 as, for example, Cell A in the 30 hours, 00 minutes frame; Cell A2 in the 34 hours, 15 minutes frame; and Cell B in the 35 hours, 10 minutes frame.
Eight digitized pictures illustrating the evolution of HNSCC cells at different moments (after the beginning of cell tracking). Two particular cells, labeled A and E, were identified at moment t = 30 hours 00 minutes. Their evolutions are shown in the following pictures over a period of 41 hours. The first generations of Cell A are labeled A1 and A2, the second generations A11, A12, A21, and A22, and similarly for cells E and B (see text for details). Dying cells are located by the CD (cell dying) label as long as they remain visible. The typical mitosis morphologies are illustrated for cell A in the 30 hours 00 minutes frame, cell A2 in the 34 hours 15 minutes frame, and cell B in the 35 hours 10 minutes frame. By the way of an example, starting from their appearance in the 44 hours 10 minutes frame, the trajectories of cells E11 and E12 are shown in the last frame (covering a period of about 27 hours).
The effects of S100A2 inhibition (by means of antisense oligonucleotides) on FADU cell migration were analyzed first of all. In accordance with the results reported in Figure 3A and B, a strong antisense effect appeared at cell motility level during the first 12 hours of cell tracking. Indeed, Figure 5A shows that a specific stimulation of about 40% was observed in terms of the greatest linear distance covered (maximum relative distance to the origin, MRDO) by the antisense-treated cells in relation to the control (CT) and CL-A2 conditions during the first 12 hours of tracking (Fig. 5B: approximately 180 trajectories analyzed per condition and pooled from three independent experiments). This effect progressively disappeared over the following hours (data not shown). In the matter of cell speed, a clear stimulatory effect (about 20%) manifested itself for the antisense-treated cells between the 12th and 36th hour after the beginning of tracking (Fig. 5B: between 250 and 350 trajectories analyzed per condition) and disappeared during the final 12 hours of tracking (data not shown).
Illustration of the effects (mean and sem) of oligonucleotides (0.1μm) on FADU cell motility. Panel A reports the greatest linear distance covered (MRDO) by cells over the first 12 hours of tracking (about 180 measurements per condition, pooled from 3 independent experiments), and Panel B the average cell speed, evaluated over 24 hours from the 12th to the 36th hours of tracking (between 250 and 350 measurements per condition, pooled from three independent experiments). The levels of significance (< 0.05) are also reported in comparison with control.
Secondly, the effects on RPMI cell migration of S100A2 added externally at three distinct concentrations (ie, 0.1, 1, and 10 nm) were investigated. Figure 6 illustrates the effects so produced on the greatest linear distance covered (MRDO) by the RPMI cells over 48 hours of observation (similar effects were observed in terms of the average speed, data not shown). Whereas Figure 6C reports the effects obtained (over 48 hours) if all the tracked cells are taken into account (between 500 and 700 trajectories per condition), Figure 6A and B, shows the effects observed on cell subpopulations consisting of the 25% and 50% most motile cells selected in each experimental condition (this was effected by comparing the 25% to 50% highest MRDO values selected in each condition). These results show that the three S100A2 concentrations very significantly reduced the greatest distance covered by the half-population made up of 50% of the most motile RPMI cells, whereas only the 10 nm concentration statistically reduced the greatest distance covered by 100% of the cells analyzed. In fact, more detailed analyses indicated that the S100A2-induced effects reported above were particularly stable between the 12th and 36th hours of tracking (data not shown).
The effects (mean and sem) of externally added S100A2 on the greatest linear distance covered (MRDO) by the RPMI cells over 48 hours of observation. This figure reports the results obtained by comparing the x% of highest MRDO values (ie, the most motile cells for x = 25, 50, or 100) obtained in each treated condition with those in control (100% represents between 500 and 700 trajectories per condition pooled from 3 independent experiments).
As described in “Materials and Methods,” our cell-tracking device also enabled us to evaluate different parameters related to cell kinetics (ie, division, death, overall growth, and cell-cycle time) and so to check the possible effects on cell viability of the different cell treatments used in the migration studies (ie, 0.1μm oligonucleotides or externally added S100A2) over 48 hours of observation. No appreciable change was observed in the cell kinetics (and thus in cell viability) in response to the different cell treatments carried out. A possible exception was the decrease in the cell-cycle time of the S100A2-treated RPMI cells, a phenomenon which only occurred at the lowest dose tested (ie, 0.1 nm, p < 0.01) (data not shown).
Influence of S100A2 on Actin Polymerization/depolymerization Dynamics in RPMI Cells
Figure 1E and F, illustrates RPMI cells cultured for 24 hours in the presence (Fig. 1F) or absence (control; Fig. 1E) of 1 nm S100A2. The glomerular actin cytoskeleton is shown in red fluorescence and the fibrillary actin cytoskeleton in green. The quantitative determination of each type of fluorescence showed that at 1 and 10 nm (but not at 0.1 nm) S100A2 markedly reduced the amount of glomerular actin regardless of whether the RPMI cells were cultured for 30 minutes (data not shown) or 24 hours (Fig. 7A) in the presence of S100A2. The data clearly indicate that externally added S100A2 markedly modifies the polymerization/depolymerization dynamics of actin and that this effect appears rapidly (within 30 minutes, data not shown) and remains stable for at least 24 hours (Fig. 7A). The marked S100A2-induced decrease in the amount of glomerular actin (red fluorescence) was the mirror image of an increase in the amount of fibrillary actin (green fluorescence) (data not shown).
A, Quantitative determination (mean, sem, and sd) of the amount of glomerular actin in RPMI cells cultured for 24 hours in the absence (0 nm) or presence of externally added S100A2 (at 0.1, 1.0, and 10.0 nm). B, Receptor for advanced glycation end-products (RAGE) expression in the five HNSCC cell lines analyzed by means of RT-PCR analysis on the same cDNA samples as those used for the analyses shown in Figure 2A.
Presence of RAGE Receptors and the Binding of S100A2 to Cells
In view of the extracellular activities of S100A2 reported above on RPMI cells, we investigated whether the so-called RAGE receptors are present on these cells. As discussed below, RAGE is the only known receptor candidate for S100/calgranulin polypeptides. Figure 7B reports the results of an RT-PCR analysis performed on the five cell lines studied. In fact, this analysis was carried out on the same cDNA samples as those used for the S100A2 RT-PCR analysis reported in Figure 2A. Interestingly enough, Figures 7B and 2A clearly show an inverse correlation between RAGE and S100A2 mRNA expression. Indeed, the two cells lines that expressed the most S100A2 (ie, FADU and SCC-25) did not express RAGE, and, inversely, the cell line which expressed the least S100A2 (ie, RPMI) expressed the most RAGE. Evidence that externally added S100A2 is able to bind to RPMI cells was provided by the fact that we observed a change in S100A2 immunofluorescence after the addition of S100A2 to the culture media of the RPMI cells, as illustrated in Figure 1G and H. Figure 1G shows untreated RPMI cells (control), for which the level of S100A2 immunofluorescence (in red) was very low in comparison with the higher level illustrated in Figure 1H and observed 30 minutes after the addition of 10 nm of S100A2 to the culture media. The red film appearing on the RPMI cells in Figure 1H can be interpreted as being due to the binding of S100A2 to RPMI cells via RAGE or another type of receptor. We performed a similar experiment with the FADU cells. However, we were not able to identify a possible change in S100A2 immunofluorescence after the addition of S100A2 to the culture media of these cells because they showed a high level of S100A2 immunofluorescence even when untreated (data not shown).
Discussion
S100 Ca2+-binding proteins have recently become of major interest because of their differential expression in neoplastic tissue, their involvement in metastatic processes, and the clustering organization of at least 13 S100 genes on human chromosome 1q21, a region frequently rearranged in several types of tumor (Ilg et al, 1996). A tumor suppressor role for S100A2 is also suggested for different reasons, such as the repressed expression in a large number of tumors, regulation by immediate early genes that are important regulators of normal growth control, and selective hypermethylation within the promoter region (a inactivation mechanism of human tumor-suppressor genes) (Wicki et al, 1997). Our recent data on epithelium tissue (N Nagy, D Hoyaux, I Gielen, BW Schafer, R Pochet, CW Heizmann, R Kiss, I Salmon, and C Decaestecker, unpublished data) support these arguments by demonstrating a decrease in S100A2 staining in epithelial tissue in cancer as opposed to normal cases. Because we are particularly interested in the invasiveness of head and neck squamous cell carcinomas (Choufani et al, 1999; Delorge et al, 2000; Saussez et al, 1998), we decided to investigate the S100A2 function in vitro as a potential growth and/or cell migration inhibitor in the case of epithelial cancer cells of squamous origin. We hypothesized that in epithelial cancer cells, S100A2 could play its tumor suppressor role at a level of cell migration that would no longer be restrained when S100A2 ceased to be expressed. Cancer cells could thus migrate anarchically toward normal tissue as a result of the turn-off of S100A2 expression in tumoral epithelial tissue.
From the five HNSCC cell lines analyzed, we selected the FADU and the RPMI models because of their different levels of S100A2 expression, as clearly confirmed by RT-PCR, Western blotting, and immunochemistry (Fig. 2). The heterogeneous S100A2 expression observed in the five cell lines had to be related to their heterogeneous origins (see “Materials and Methods”) involving various levels of cell differentiation and aggressiveness. Indeed, even in the case of a given origin (eg, laryngeal squamous cell carcinomas), Lauriola et al (2000) report that S100A2 expression can be very heterogeneous in the function of the tumor grade and the degree of squamous differentiation which, in HNSCCs, ranges from well-differentiated keratinizing to anaplastic nonkeratinizing tumors (Wiernik et al, 1991). More particularly, S100A2 expression has been associated positively with squamous-cell differentiation and negatively with tumor grading (Shrestha et al, 1998; Lauriola et al, 2000). Unfortunately, too few data are available from the American Type Culture Collection (ATCC, Manassas, Virginia) to provide a rigorous explanation of the different S100A2 expression patterns in the cell lines analyzed. However, it will be noted that although the RPMI cell line was established from a tumor diagnosed as an anaplastic squamous cell carcinoma (which may explain its low S100A2 expression level), the FADU cell line is reported as producing well-differentiated grade I epidermoid carcinomas in nude mice (which is in agreement with a high S100A2 expression level) (Hay et al, 1994).
In the present study two complementary approaches were investigated, namely, the inhibition of S100A2 expression by means of specific antisense oligonucleotides in a cell line with a high level of expression (FADU), and extracellular S100A2 treatment in a cell line with a low level of expression (RPMI) in accordance with the extracellular effects reported in the literature (as discussed later). Our data show that although S100A2 markedly modifies cell migration features, it does not affect cell kinetic features. Indeed, as is shown by the strong S100A2-induced modification observed in the RPMI model, S100A2 acts as a potent inhibitor at the cell migration level. This effect is confirmed by the antisense oligonucleotide treatment that stimulated FADU cell motility.
The cellular effector protein has yet to be determined for most members of the S100 family, but recent findings indicate that cytoskeletal elements are most likely to be involved as, indeed, has already been demonstrated for the actin-associated protein calponin (CaP), caldesmon (CaD), the nonmuscle myosin heavy chain, vimentin, the glial fibrillary acidic protein, and tubulin (Donato, 1999; Gimona et al, 1997; Schäfer and Heizmann, 1996). The S100A2-mediated role in the migration of living cancer cells, evidenced here in relation to actin polymerization/depolymerization dynamics, is thus fully corroborated by molecular-biology-related data. Because different microfilament populations are associated with different cellular compartments (such as growth cones, filipodia, stress fibers, and lamellipodia), and because these different populations are made up of specific associations between different isoforms of the two core microfilament components, ie, actin and tropomyosin (Gunning et al, 1998), all that now remains to be clarified is the specific association of actin/tropomyosin isoform(s) on which S100A2 could act. Indeed, epithelial cells express a complex pattern of tropomyosin isoforms that occupy differential sites in the cells and display different patterns of expression depending on their origins and stages of differentiation (Temm-Grove et al, 1998).
Extracellular activities have been reported for various S100 proteins (Donato, 1999; Schäfer and Heizmann, 1996). More particularly, Komada et al (1996) report that S100A2 is released by cultured LLC-PK1 cells and interacts with eosinophil receptors, thus producing an intracellular Ca2+ response. However, the biochemical nature of these S100 receptor proteins has not yet been elucidated. A recent advance in this matter was made by Hofmann et al (1999), who showed that S100/calgranulin polypeptides bind to RAGE. When secreted, S100A2 might thus signal via RAGE. It is nevertheless interesting to note that the S100A2 (Glenney et al, 1989) and the RAGE (Neeper et al, 1992; Schmidt et al, 1992) were both isolated from bovine lungs. In the present study, we found evidence of a surprisingly negative association between the presence of RAGE in the cell lines and their levels of S100A2 expression. In particular, the RPMI cells, for which we demonstrated extracellular S100A2 effects, appeared to be relatively rich in RAGE mRNAs. Concerning secretion, because S100 proteins lack the structural requirements for the standard endoplasmic reticulum/Golgi route, the process of secretion is also unclear. However, a microtubule-dependent pathway has been demonstrated by Rammes et al (1997).
In conclusion, the data from the present study show that S100A2 clearly modifies the cell motility features in the FADU and RPMI cell lines (while only slightly modifying the cell kinetic parameters). S100A2, for which a putative role as a tumor suppressor gene has been advocated, therefore seems to exert a particular antitumoral influence by restraining the cell migration potential of epithelial cancer cells, or at least those of squamous origins. This influence on cancer cell migration relates, at least in part, to marked S100A2-induced modifications at the level of actin polymerization/depolymerization dynamics.
Materials and Methods
Cell Lines and Culture Media
Five human HNSCC cell lines from the American Type Culture Collection (ATCC) were used. These were the FADU (originating from a hypopharynx, code HTB-43), the Det-562 (from a pharynx, code CCL-138), the RPMI (from a nasal septum, code CCL-30), the SCC-9 (from a tongue, code CRL-1629), and the SCC-25 (from a tongue, code CCL-1628) cell lines. The cells were cultured at 37° C in sealed (air-tight) Falcon plastic dishes (Nunc; Gibco, Merelbeke, Belgium) containing Eagle’s minimal essential medium (MEM; Gibco), supplemented with 10% fetal calf serum (FCS). All the media were supplemented with a mixture of 0.6 mg/ml glutamine (Gibco), 200 IU/ml penicillin (Gibco), 200 IU/ml streptomycin (Gibco), and 0.1 mg/ml gentamycin (Gibco). The FCS was heat-inactivated for 1 hour at 56° C. Some posttreatments (such as oligonucleotide incubation) requiring changes in the culture media are detailed in the sections that follow.
Immunohistochemical Staining
Returning to the five cell lines cultured, we made cell pellets by centrifuging 10 million cells from each cell line for 10 minutes at 800 × g. These pellets were then fixed for 4 days in buffered formalin (4%), dehydrated, and embedded in paraffin wax. Three pellets were available for each of the five lines. Two sections of 5 μm each were taken from each pellet and clinical sample and subjected to processing with the anti-S100A2 antibody and kit reagents, as previously described (Camby et al, 1999). Briefly, incubation with the antibody against human S100A2 protein was carried out at 25 ± 1° C for 60 minutes. The dilution used was 1:500. Immunoreaction was developed by applying the avidin-biotin-peroxidase complex (ABC) kit reagents (Vector Labs, Burlingame, California), with diaminobenzidine/H2O2 as the chromogenic substrates (Goldschmidt et al, 1996; Jannot et al, 1993). The control tissues were incubated either with the corresponding pre-immune sera and the second antibody alone, or with polyclonal anti-S100A2 antibody preabsorbed with human recombinant S100A2. Counterstaining was performed with hematoxylin. The antibody against human recombinant S100A2 was raised in rabbits. Its characteristics have been described earlier (Huang et al, 1996; Ilg et al, 1996).
Computer-Assisted Microscopy for Quantitative Immunochemistry on the Five HNSCC Cell Lines
With a view to characterizing the S100A2 immunohistochemical expression in the HNSCC cell lines, two quantitative variables were determined by means of a SAMBA 2005 computer-assisted microscope system (UNILOG, Grenoble, France) with a 40 × magnification lens (BX50 microscope, aperture 0.65; Olympus, Tokyo, Japan). The labeling index (LI) refers to the percentage of cells specifically stained by a given histochemical marker. The mean optical density (MOD) denotes staining intensity. Ten fields of between 60,000 and 120,000 μm2 each were scanned for each cell line. The way in which we used the computer-assisted system to quantify the staining is detailed elsewhere, as are all the standardization procedures dealing with the manner in which we used the computer-assisted microscopy (Choufani et al, 1999; Goldschmidt et al, 1996; Saussez et al, 1998). A negative histological control slide (from which the primary anti-S100A2 antibody was omitted) was analyzed for each specimen under study. The software used on the computer-assisted microscope automatically subtracted the LI and MOD values of the negative control samples from each of the corresponding positive ones.
RT-PCR Analyses
The cells were grown to subconfluence in Petri dishes and were lysed by the addition of TRIPURE isolation reagent (Roche Diagnostics, Mannheim, Germany). Total RNA was prepared according to the manufacturer’s recommendation, and 5 μg RNA was used as a template for cDNA synthesis. Reverse transcription was performed for 60 minutes at 42° C in First Strand buffer (50 mm Tris/HCl, pH 8.3; 75 mm KCl; 3 mm MgCl2), 10 mm dithiothreitol (DTT), deoxyribonucleoside triphosphates (dNTPs) (0.5 mm each), random hexamer primers (100 ng/μl), and 200 U SuperscriptII reverse transcriptase (Gibco BRL, Merelbeke, Belgium). The reaction was terminated by incubation for 15 minutes at 72° C. The integrity of the cDNA and the homogeneity of the samples were confirmed by a glyceraldehyde-3-phosphate dehydrogenase (GADPH)-specific PCR analysis. The primers used for RT-PCR analysis were designed by using the HYBsimulator program (Advanced Gene Computing Technologies, Irvine, California) (Hyndman et al, 1996). They were as follows: S100A2 sense, 5′-CACTACCTTCCACAAGTACT-3′; and antisense, 5′-GAAGTCATTGCACATGACAG-3′; RAGE sense, 5′-CCTAATGAGAAGGGAGTATCTG-3′; and antisense, 5′-CACAAGATGACCCCAATGA-3′ (purchased from Biosource Europe S.A., Nivelles, Belgium). All the PCR analyses were carried out in 50 μl reaction media containing 2 μl cDNA, 2.5 U Taq polymerase and PCR buffer (Gibco BRL), dNTPs (25 μm each), and 0.17 μm of both sense and antisense primers. Amplification was performed as follows: 94° C for 4 minutes, followed by 35 (for S100A2 and RAGE) or 30 (for GADPH) cycles at 94° C for 45 seconds, 55° C for 45 seconds, and 72° C for 45 seconds. After a final extension period of 10 minutes at 72° C, the amplified products were separated by gel electrophoresis in 2% TAE gels and visualized under UV light by means of ethidium bromide staining.
Western Blotting Analyses
Cell extracts were prepared by the sonication of scrapped subconfluent cells in PBS containing 1 mm phenylmethylsulfonyl fluoride (PMSF) and 10 μg/ml aprotinin. After sonication, the protein concentrations were normalized (BCA protein assay; Pierce, distributed by Polylabo, Antwerp, Belgium) and 30 μg loaded in each lane of a 12% polyacrylamide gel under denaturing and reductive conditions. After electrophoresis, the proteins were transferred onto a Polyscreen polyvinylidene difluoride (PVDF) membrane (NEN Life Science Products, Boston, Massachusetts). After the blockage of nonspecific binding, the filters were incubated with S100A2 primary antibody (dilution 1/250 in 0.1% Tween 20 in Tris buffered saline [T-TBS] containing 5% of milk powder) overnight at 4° C. The filters were then washed and placed for 1 hour at room temperature in T-TBS containing 5% of milk powder and a horseradish peroxidase-conjugated secondary antibody (dilution 1/5000). The visualization of the immunoreactive proteins was carried out with an enhanced chemiluminescence reaction (NEL103, NEN) by exposing the membranes to hyperfilm electrochemiluminescence (ECL) (Amersham Life Science, Buckinghamshire, United Kingdom). Control experiments included the omission of the incubation step with the specific antiprotein antibody as the negative control, and the electrophoresis of 1 ng/ml of recombinant S100A2 as the positive one. The molecular weight of S100A2 is 11kD.
Antisense oligonucleotides
S100A2-specific antisense (AS-A2) and random control (CL-A2) oligonucleotides were also designed by using the HYBsimulator program (Advanced Gene Computing Technologies, Irvine, CA) (Hyndman et al, 1996; Mitsuhashi, 1997). The 15-mer sequences detected in this way were: AS-A2 = 5′-GGAAAACAGCATACT-3′ and CL-A2 = 5′-TCATACGACAAGAGA-3′. Phosphorothioate oligonucleotides labeled (in 5′) with fluorescein isothiocyanate (FITC) were purchased from Biosource Europe S.A. (Nivelles, Belgium). For the immunofluorescence and cell tracking experiments (see below) the FADU cells were seeded in 24-well plates (immunofluorescence) or 4-well plates (tracking) for 3 days in conventional media (detailed above). After the cells had been washed, the media were replaced by serum-free media in the presence/absence of oligonucleotides at different concentrations (0.1, 1 and 10 μm). The cells were then incubated for different periods (12 hours, 24 hours and 48 hours). The incubation with oligonucleotides was performed 1 hour before the beginning of the cell tracking.
Human Recombinant S100A2
We analyzed the effects of externally added S100A2 on migration features and actin polymerization/depolymerization dynamics in the RPMI cells (see below). For this purpose we used human recombinant S100A2 protein. The purity and biochemical properties of this S100A2 protein have been described elsewhere (Franz et al, 1998; Stradal et al., 2000).
Immunofluorescence Assays
Immunofluorescence was first used to verify antisense oligonucleotide specificity. This was done by quantifying the S100A2 expression in FADU cells, either left untreated or treated with oligonucleotides (AS-A2 or CL-A2). After 12 hours or 24 hours of treatment (see above), the cells were washed thoroughly in cold phosphate buffer (PBS, pH 7.4) to eliminate extracellular oligonucleotides before fixation for 20 minutes in 4% buffered-formol at 4° C. After three washes in PBS, the cells were gently permeabilized for 1 minute at 4° C with PBS containing 2 mm MgCl2, 0.2 mm dithiothreitol, 10% glycerol, and 0.01% Triton × 100. After three washes in PBS, the cells were blocked with 1% horse serum (v/v in PBS) (Vector Laboratories, Burlingame, California) for 20 minutes before being incubated for 45 minutes at room temperature with the rabbit anti-human S100A2 antibody at a 1/500 dilution in PBS containing 0.1% BSA. After three washes in PBS, the coverslips were incubated for 45 minutes in a dark room with a mixture of Alexa 594 (red) conjugated anti-rabbit IgG (1/400; Molecular Probes, Eugene, Oregon) and Alexa 488 (green) conjugated anti-fluorescein IgG (1/400, Molecular Probes) in PBS containing 0.1% BSA. After three washes in PBS, the coverslips were mounted with Vectashield (Vector Laboratories). Photographs of between 60 and 100 cells per experimental condition (pooled from three independent experiments) were made with a × 100 lens in the same carefully checked exposure conditions and submitted to quantification (see below).
We proceeded in the same way to investigate whether externally added S100A2 (10 nm) was able to bind RPMI cells. However, in this case, the cells were not permeabilized and the coverslips were incubated with a mixture of Dapi (Molecular Probes) (to identify cell nuclei) and Alexa 594 (red) conjugated anti-rabbit IgG (1/400).
Finally, the influence of the S100A2 protein on the actin assembly in the RPMI cells was investigated by means of fluorescent probes specific to either filamentous (F) or monomeric (G) actin. The fluorescent phallotoxin derivative phallacidin was chosen to label the F-actin, and fluorescent DNaseI to label the G-actin (Knowles and McCulloch, 1992). Both compounds bind to actin subunits in a stoichiometric ratio (Knowles and McCulloch, 1992). The experimental protocol was as follows. RPMI cell concentrations of 40,000 cells/ml MEM were plated for 48 hours on glass histological slides before being treated or not treated (control) with 0.1, 1, or 10 nm S100A2. After 24 hours of treatment, the cells were washed, fixed, and permeabilized as explained above. After three rinses in PBS, the cells were incubated for 30 minutes at 20° C with DNaseI conjugated with Texas Red (DNaseI-TR, Molecular Probes) at a final concentration of 10 μg/ml PBS. The cells were then washed three times in PBS before undergoing incubation for 30 minutes at 20° C with phallacidin conjugated with the fluorescein-like fluorochrome BODIPY FL (Phall-FL, Molecular Probes) at a final concentration of 5 units/ml PBS. Methanol from the phallacidin stock solution was evaporated before dilution in PBS. After three to five rinses in PBS, the slides were mounted with Vectashield mounting medium (Vector Laboratories). Photographs of 200 cells per experimental condition were made (for both fluorochromes) with a × 40 lens in the same carefully checked exposure conditions.
The fluorescent signals were observed and quantified by means of a Zeiss Axioplan microscope (Zeiss, Jena, Germany) equipped with excitation/emission filters (590/615 nm for red fluorochromes and 505/512 for green ones) and a Hamamatsu C4880 Dual-mode cooled CCD camera (Hamamatsu Phototronics, Hamamatsu, Japan) linked to a software developed by SAMBA Technologies (Grenoble, France).
Immunoprecipitation Assay
This assay was used to demonstrate the oligonucleotide effects on the neosynthesis of the S100A2 protein in the FADU cells. After 1 hour’s incubation in methionine and cysteine-free medium (DMEM; ICN Biomedicals, Irvine, California), the FADU cells were either left untreated or treated with oligonucleotides (AS-A2 or CL-A2) at 0.01 and 0.1 μm for over 12 hours. Five hours before the end of this treatment the cells were labeled metabolically with 0.1 mCi/ml of TRANS35S-LABEL (1175 Ci/mmol, ICN) in DMEM. Cell extracts were obtained with a RIPA lysis buffer (0.1M Tris HCl, pH 7.4; 0.15 M NaCl; 1 mm PMSF; 0.1% SDS; 1 mm EDTA; 1% TRITON X-100; 0.5% aprotinin; and 0.5% sodium deoxycholate [Cheresh and Spiro, 1987]) and centrifuged at 3000 × g for 30 minutes at 4° C. Supernatants were loaded onto 40 μl of Protein G Sepharose 4 Fast Flow (Pharmacia Biotech, Uppsala, Sweden). The anti-S100A2 antibody (1/200) was added and incubated for 2 hours at 4° C. Precipitated immune complexes were washed successively with phosphate buffer (PBS) containing 0.1% Triton X-100, PBS containing 0.5 mm NaCl, and PBS alone before being diluted in Laemmi buffer (100 mm Tris-HCl, pH 6.8; 4% SDS; 0.2% Bromophenol blue; 20% glycerol). Samples were analyzed by electrophoresis on 15% SDS PAGE. Radiolabeled immunoprecipitated proteins were visualized by autoradiography. The negative control involved the omission of the antibody to check the nonspecific labeling. The levels of immunoprecipitated protein were quantitatively determined by means of a gel scanner and Bioprofil Image Analysis Software (Proxilab, Bordeaux, France).
Cell Tracking Procedure
As detailed elsewhere (DeHauwer et al, 1997, 1998, 1999), we have developed a prototype able to quantitatively determine the migration and kinetic features of living cells. This device enables living cells cultured in Falcon dishes containing a buffered medium to be maintained in a closed environment at a thermostat setting of 37.0 ± 0.1° C for the time required for an experiment (eg, from a few minutes to several days) (DeHauwer et al, 1997, 1999). The living cells are recorded via a CCD camera placed on a phase-contrast microscope. A control monitor placed between the camera and the computer enables what is happening in the culture dishes to be visualized at all times. Specialized software has been developed to control this system and analyze the data recorded (DeHauwer et al, 1999), as illustrated in Figure 4. Digitized images are in fact recorded every 4 minutes for a given period and enable cell evolution to be followed in terms of both cell kinetics (division, death, etc.) and migration.
In the present experiments, we first investigated the biological behavior (over 48 hours) of FADU cells, either left untreated or treated with either the S100A2 antisense oligonucleotide (AS-A2) or its random version (CL-A2) at 0.1 μm (in accordance with immunofluorescence and immunoprecipation results). We then recorded (over 48 hours) the behavior of RPMI cells, either treated with S100A2 or left untreated. The S100A2-induced influence was analyzed at three different doses of 0.1, 1, and 10 nm. The treatments with oligonucleotides or S100A2 were carried out 1 hour before the beginning of cell tracking. Each experimental condition was performed independently and in triplicate.
Because our system is sufficiently efficient to distinguish between cell deaths and mitoses on the basis of morphological criteria (Fig. 4), suitably adapted software applications included in this system enabled us to quantify the number of mitoses produced during the entire experiment (in the form of the number of cells present during the experiment) in addition to the migration variable described below. This quantification refers to the rate of proliferation, ie, to the proliferating cell fraction. Also, by using the experimental method described above, we were able to evaluate the rate of proliferation, ie, the length of the cell cycle, by determining the number of minutes that elapsed between each consecutive mitosis from a given “ancestor” cell. The overall growth of the cell population analyzed was established by dividing the number of cells present at time t = 48 hours 00 minutes (ie, the number of cells present at the end of the experiment) by the number of cells present at time t = 0 minutes (the beginning of the cell tracking).
Cell migration was determined as detailed elsewhere (DeHauwer et al, 1997, 1998, 1999). Briefly, our software enabled each cell in the culture under study to be identified against the background (in the present case, the plastic Falcon dishes) and labeled by the coordinates of its centroid. Because this operation was performed for every frame of the sequence (every 4 minutes), the trajectory of each cell centroid could be computed by interpolation, as illustrated in the case of cells E11 and E12 in the last frame of Figure 4. Each trajectory was then characterized by two quantitative motility descriptors, namely, the average speed and the “maximum relative distance to the origin” (the MRDO variable) of each cell. This latter variable was the greatest linear distance found between a cell’s original and its subsequent positions normalized by the observation time of the cell analyzed (DeHauwer et al, 1997, 1998, and 1999). Between 20 and 30 cells were present at the beginning of each of the three experiments performed for a given experimental condition. By the end of the 48 hours of cell tracking, between 100 and 300 trajectories had been analyzed per experiment. The software used also enabled us to select and analyze specific periods (eg, of 12 hours).
Statistical Analyses
The statistical comparisons of the quantitative data were carried out by means of the Student t tests after a check of the equality of variance by means of the Levene test, and by the normal distribution fitting of the data by means of the χ2 test of goodness of fit. When these parametric conditions were not satisfied, the nonparametric Mann-Whitney tests were applied. All the statistical analyses were carried out using Statistica (Statsoft, Tulsa, Oklahoma).
References
Camby I, Nagy N, Lopes MB, Schäfer BW, Maurage CA, Ruchoux MM, Murmann P, Pochet R, Heizmann C, Brotchi J, Salmon I, Kiss R, and Decaestecker C (1999). Supratentorial pilocytic astrocytomas, astrocytomas, anaplastic astrocytomas and glioblastomas are characterized by a differential expression of S100 proteins. Brain Pathol 9: 825–843.
Cheresh DA and Spiro RC (1987). Biosynthetic and functional properties of an Arg-Gly-Asp-directed receptor involved in human melanoma cell attachment to vitronectin, fibrinogen, and von Willebrand factor. J Biol Chem 262: 17703–17711.
Choufani G, Saussez S, Marchant H, Bishop P, Schüring MP, Danguy A, Salmon I, Gabius HJ, Kiss R, and Hassid S (1999). Galectin-1, galectin-3 and the Thomsen-Friedenreich antigen expressions decrease as clinical aggressiveness increases in head and neck cancers. Cancer 86: 2353–2363.
DeHauwer C, Camby I, Darro F, Decaestecker C, Gras T, Salmon I, Kiss R, and VanHam P (1997). Dynamic characterization of glioblastoma cell motility. Biochem Biophys Res Commun 232: 267–272.
DeHauwer C, Camby I, Darro F, Migeotte I, Decaestecker C, Verbeek C, Danguy A, Brotchi J, Salmon I, Van Ham P, and Kiss R (1998). Gastrin inhibits motility, decreases cell death and increases cell proliferation in human glioblastoma cell lines. J Neurobiol 37: 373–382.
DeHauwer C, Darro F, Camby I, Kiss R, Van Ham P, and Decaestecker C (1999). In Vitro motility evaluation of aggregated cancer cells by means of automatic image processing. Cytometry 36: 1–10.
Delorge S, Saussez S, Pelc P, Devroede B, Marchant H, Schüring MP, Danguy A, Salmon I, Gabius HJ, Kiss R, and Hassid S (2000). Correlation of galectin-3/galectin-3-binding sites with low differentiation status in head and neck squamous cell carcinomas. Otolaryngol Head Neck Surg 122: 834–841.
Donato R (1999). Functional roles of S100 proteins, calcium-binding proteins of the EF-hand type. Biochim Biophys Acta 1450: 191–231.
Franz C, Durussel I, Cox JA, Schafer BW, and Heizmann CW (1998). Binding of Ca2+ and Zn2+ to human nuclear S100A2 and mutant proteins. J Biol Chem 273: 18826–18834.
Gimona A, Lando Z, Dolginov Y, Vandekerckhove J, Kobayashi R, Sobieszek A, and Helfman DM (1997). Ca2+-dependent interactions of S100A2 with muscle and nonmuscle tropomyosins. J Cell Sci 110: 611–621.
Glenney JR Jr, Kindy MS, and Zokas L (1989). Isolation of a new member of the S100 protein family: Amino acid sequence, tissue, and subcellular distribution. J Cell Biol 108: 569–578.
Goldschmidt D, Decaestecker C, Berthe JV, Gordower L, Remmelink M, Danguy A, Pasteels JL, Salmon I, and Kiss R (1996). The contribution of computer-assisted methods for histopathological classification of adipose tumors. Lab Invest 75: 295–306.
Gunning P, Hardeman E, Jeffrey P, and Weinberger R (1998). Creating intracellular structural domains: Spatial segregation of actin and tropomyosin isoforms in neurons. Bioessays 20: 892–900.
Hay R, Caputo J, Chen TR, Macy M, McClintock P, and Reid Y, editors (1994). ATCC cell lines and hybridomas, 8th ed. Rockville, MD: American Type Culture Collection, 15, 260.
Heizmann CW and Cox JA (1998). New perspectives on S100 proteins: A multi-functional Ca(2+), Zn(2+)- and Cu(2+)-binding protein family. Biometals 11: 383–397.
Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, and Schmidt AM (1999). RAGE mediates a novel proinflammatory axis: A central cell surface receptor for S100/calgranulin polypeptides. Cell 97: 889–901.
Huang JW, Ming Z, Shrestha P, Mori M, Ilg E, Schäfer BW, and Heizmann CW (1996). Immunohistochemical evaluation of the Ca2+-binding S100 proteins S100A1, S100A2, S100A4, S100A6 and S100B in salivary gland tumors. J Oral Pathol Med 25: 547–555.
Hyndman D, Cooper A, Pruzinsky S, Coad D, and Mitsuhashi M (1996). Software to determine optimal oligonucleotide sequences based on hybridization simulation data. Biotechniques 20: 1090–1097.
Ilg EC, Schäfer BW, and Heizmann CW (1996). Expression pattern of S100 calcium-binding proteins in human tumors. Int J Cancer 68: 325–332.
Jannot MC, Kruczynski A, Limouzy A, Dangou JM, Selves J, Delsol G, Martinez J, and Kiss R (1993). Spontaneous evolution of cytoplasmic lectin binding and nuclear size and deoxyribonucleic acid content in human colorectal cancer grafted onto nude mice. Lab Invest 68: 446–455.
Knowles GC and McCulloch CAG (1992). Simultaneous localization and quantification of relative G and F actin content: Optimization of fluorescence labeling methods. J Histochem Cytochem 40: 1605–1612.
Komada T, Araki R, Nakatani K, Yada I, Naka M, and Tanaka T (1996). Novel specific chemtactic receptor for S100L protein on guinea pig eosinophils. Biochem Biophys Res Commun 220: 871–974.
Lauriola L ., Michetti F, Maggiano N, Galli J, Cadoni G, Schäfer BW, Heizmann CW, and Raneletti FO (2000). Prognostic significance of the Ca2+ binding protein S100A2 in laryngeal squamous-cell carcinoma. Int J Cancer 89: 345–349.
Lee SW, Tomasetto C, and Sager R (1991). Positive selection of candidate tumor-suppressor genes by substractive hybridization. Proc Natl Acad Sci USA 88: 2825–2829.
Lee SW, Tomasetto C, Swisshelm K, Keyomarsi K, and Sager R (1992). Down-regulation of a member of the S100 gene family in mammary carcinoma cells and reexpression by azadeoxycytidine treatment. Proc Natl Acad Sci USA 89: 2504–2508.
Maelandsmo GM, Florenes VA, Mellingsaeter T, Hovig E, Kerbel RS, and Fodstad O (1997). Differential expression patterns of S100A2, S100A4 and S100A6 during progression of human malignant melanoma. Int J cancer 74: 464–469.
Mitsuhashi M (1997). Strategy for designing antisense oligonucleotide sequences. J Gastroenterol 32: 282–287.
Moore B (1965). A soluble protein characteristic of the nervous system. Biochem Biophys Res Commun 19: 739–744.
Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, and Shaw A (1992). Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem 267: 14998–15004.
Pedrocchi M, Schafer BW, Mueller H, Eppenberger U, and Heizmann CW (1994). Expression of Ca(2+)-binding proteins of the S100 family in malignant human breast-cancer cell lines and biopsy samples. Int J Cancer 57: 684–690.
Rammes A, Roth J, Goebeler M, Klempt M, Hartmann M, and Sorg C (1997). Myeloid-related protein (MRP) 8 and MRP14, calcium-binding proteins of the S100 family, are secreted by activated monocytes via a novel, tubulin-dependent pathway. J Biol Chem 272: 9496–9502.
Saussez S, Marchant H, Nagy N, Decaestecker C, Hassid S, Jortay A, Schüring M, Gabius HJ, Danguy A, Salmon I, and Kiss R (1998). Quantitative glycohistochemistry defines new prognostic markers in cancers of the oral cavity. Cancer 82: 252–260.
Schäfer BW and Heizmann CW (1996). The S100 family of EF-hand calcium-binding proteins: Functions and pathology. Trends Biochem Sci 21: 134–140.
Schmidt AM, Vianna M, Gerlach M, Brett J, Ryan J, Kao J, Esposito C, Hegarty H, Hurley W, and Clauss M (1992). Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J Biol Chem 267: 14987–14997.
Shrestha P, Muramatsu Y, Kudeken W, Mori M, Takai Y, Ilg EC, Schäfer BW, and Heizmann CW (1998). Localization of Ca2+-binding S100 proteins in epithelial tumors of the skin. Virchows Arch 432: 53–59.
Stoll SW, Zhao X, and Elder JT (1998). EGF stimulates transcription of CaN19 (S100A2) in HaCaT keratinocytes. J Invest Dermatol 111: 1092–1097.
Stradal TB, Troxler H, Heizmann CW, and Gimona M (2000). Mapping the zinc ligands of S100A2 by site-directed mutagenesis. J Biol Chem 275: 13219–13227.
Temm-Grove CJ, Jockusch BM, Weinberger RP, Schevzov G, and Helfman DM (1998). Distinct localizations of tropomyosin isoforms in LLC PK1 epithelial cells suggests specialized function at cell-cell adhesions. Cell Motil Cytoskeleton 40: 393–407.
Therior JA and Mitchison TJ (1991). Actin microfilament dynamics in locomoting cells. Nature 352: 126–131.
Wicki R, Franz C, Scholl FA, Heizmann CW, and Schäfer BW (1997). Repression of the candidate tumor suppressor gene S100A2 in breast cancer is mediated by site-specific hypermethylation. Cell Calcium 22: 243–254.
Wiernik G, Millard PR, and Haybittle JL (1991). The predictive value of histological classification into degrees of differentiation of squamous cell carcinoma of the larynx and hypopharynx compared with the survival of patients. Histopathology 19: 411–417.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nagy, N., Brenner, C., Markadieu, N. et al. S100A2, a Putative Tumor Suppressor Gene, Regulates In Vitro Squamous Cell Carcinoma Migration. Lab Invest 81, 599–612 (2001). https://doi.org/10.1038/labinvest.3780269
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780269
This article is cited by
-
The p53-S100A2 Positive Feedback Loop Negatively Regulates Epithelialization in Cutaneous Wound Healing
Scientific Reports (2018)
-
Clinical significance of S100A2 expression in gastric cancer
Tumor Biology (2014)
-
Joining S100 proteins and migration: for better or for worse, in sickness and in health
Cellular and Molecular Life Sciences (2014)
-
S100A2 protein and non-small cell lung cancer. The dual role concept
Tumor Biology (2014)
-
S100A2 in cancerogenesis: a friend or a foe?
Amino Acids (2011)