Abstract
Using comparative genomic hybridization (CGH), we have previously demonstrated frequent loss of 8p, especially its distal part, in ovarian carcinoma. To compare the deletion map of distal 8p in serous and mucinous ovarian carcinomas, we performed allelic analysis with 18 polymorphic microsatellite markers at 8p21–p23. In serous carcinoma, loss of heterozygosity (LOH) was detected in 67% of the samples, and the majority of the carcinomas showed loss of all or most of the informative markers. In contrast, only 21% of mucinous carcinomas showed allelic loss, with only one or two loci showing LOH in each sample. In serous carcinomas, LOH was associated with higher grade tumors. Three distinct minimal common regions of loss could be defined in serous carcinomas (at 8p21.1, 8p22–p23.1, and 8p23.1). Expression of a transcription factor gene, GATA4, located at one of these regions (8p23.1) was studied in serous and mucinous ovarian carcinomas by Northern blotting and immunohistochemical staining of tumor microarray. Expression was found to be lost in most serous carcinomas but retained in the majority of mucinous carcinomas. Our results suggest distinct pathogenetic pathways in serous and mucinous ovarian carcinomas and the presence of more than one tumor suppressor gene at 8p involved in the tumorigenesis of serous carcinoma.
Similar content being viewed by others
Introduction
Ovarian carcinoma originates in the gonadal surface epithelium and occurs in various forms including serous, mucinous, endometrioid, and clear cell histologies. Serous carcinoma is the most common form, comprising about 55% of ovarian carcinomas, whereas mucinous and endometrioid types account for about 10% to 15% each. All subtypes manifest as adnexal masses and currently their diagnosis, treatment, and follow-up is carried out according to the same principles. However, the risk factors and biological behavior of the various subtypes differ in many respects (Makar et al, 1995; Omura et al, 1991; Risch et al, 1996), suggesting that they could be distinct entities. This view is further supported by molecular data indicating that distinct genetic events are involved in the development of different histological subtypes of ovarian carcinoma. For example, K-ras mutations are frequent in mucinous carcinomas but are uncommon in serous carcinomas (Enomoto et al, 1991). Mutations of p53 are typical of serous carcinomas, but rare in other histological subtypes (Milner et al, 1993). Frequent PTEN/MMAC mutations have been found in endometrioid carcinomas but not in serous or mucinous ovarian carcinomas (Obata et al, 1998).
Loss of genetic material at several chromosomal regions, as detected by karyotyping, comparative genomic hybridization (CGH), and allele analysis, is frequently observed in ovarian carcinoma, suggesting the involvement of multiple tumor suppressor genes in ovarian carcinogenesis (Cliby et al, 1993; Jenkins et al, 1993; Pejovic et al, 1992; Pere et al, 1998; Sato et al, 1991; Tapper et al 1997; Yang-Feng et al, 1993). However, remarkably few tumor suppressor genes have turned out to be defective in ovarian carcinoma (Milner et al, 1993; Obata et al, 1998). The short arm of chromosome 8 is a site of frequent allelic loss in ovarian as well as in other carcinomas (Adélaide et al, 1998; Brown et al, 1999; Cliby et al, 1993; Farrington et al, 1996; Sato et al, 1991; Vocke et al, 1996; Wistuba et al, 1999; Wright et al, 1998; Yang-Feng et al, 1993), but so far no consistent tumor suppressor gene has been identified there.
The aim of this study was twofold. First, to address the suggested molecular dissimilarity of ovarian carcinoma subtypes, we compared the allelotypes of serous and mucinous carcinomas at 8p21–p23, the site of frequent loss of genetic material in ovarian carcinoma as judged by CGH analysis (Pere et al, 1998; Tapper et al 1997). When needed, microdissection was used to obtain carcinoma cell-specific DNA for the analysis. Second, because loss of heterozygosity (LOH) at distal 8p was almost exclusively detected in the serous subtype, the analysis was extended on a total of 62 serous carcinomas to define the putative tumor suppressor locus/loci more precisely. Three minimal common regions of loss were identified in serous carcinoma. The expression profile of the transcription factor gene GATA4, residing in one of them, was studied in serous and mucinous ovarian carcinomas by Northern blotting and immunohistochemical staining of tumor microarray.
Results
LOH at 8p in Serous versus Mucinous Ovarian Carcinomas
To compare the deletion map of the distal part of chromosome 8p in serous and mucinous ovarian carcinoma, we performed allelic analysis using 18 polymorphic microsatellite markers at 8p21–p23. The samples were informative on average at 13 loci (range 9–17). Of the 61 serous carcinomas included in the study, 41 (67%) showed LOH at one or more loci on 8p, and half of them showed LOH at all informative markers. In mucinous carcinomas, the pattern of LOH was clearly different. Only 3 of 14 (21%) mucinous carcinomas showed LOH, each at one or two loci: Sample 249 at marker D8S1140, Sample 783 at D8S1721, and Sample 792 at D8S552 and D8S560. The difference in the number of cases showing LOH in serous versus in mucinous carcinomas was statistically significant (p = 0.0025). The mean percentage of LOH of informative alleles in serous carcinomas was 50%, and in mucinous carcinomas it was 1.9% (p = 0.0008). Mucinous carcinomas were mostly Grade 1 and 2, whereas half of the serous carcinomas were Grade 3 (see “Materials and Methods”), which difference is due to the inherent biology of these two subtypes. When only Grade 1 and Grade 2 tumors were taken into account, the difference in the percentage of LOH of informative alleles still remained significant (37% vs 2.2%; p = 0.029). Also the stages of the serous carcinomas were higher than of the mucinous tumors. However, when only Stage I and Stage II tumors were analyzed, the difference between the groups still remained significant (53% vs 2.1%; p = 0.0016).
Microsatellite instability was detected in four serous tumor samples: in Cases 852, 223, and 412 only at one marker and in Case 210 at several markers. Because of a possible mismatch-repair-system deficiency, Case 210 was excluded from the analysis. In mucinous carcinomas one tumor sample (Case 783) presented with one microsatellite instability-positive allele.
Three Minimal Common Regions of Loss in Serous Carcinoma
Because LOH at 8p was mainly detected in serous carcinomas, we analyzed a total of 62 serous carcinoma samples to define the supposed tumor suppressor locus or loci involved in the genesis of serous ovarian carcinoma. Of these carcinomas, 21 samples (34%) showed LOH at all the informative loci and were not useful for more detailed deletion mapping. The results of LOH analyses in the 20 serous carcinomas (30%) showing partial or interstitial deletions are summarized in Figure 1A. Three distinct minimal common regions of deletion could be defined: R1 around marker D8S499, between D8S1810 and D8S1771 (8p21.1), R2 around marker D8S552, between D8S1731 and D8S640 (8p22-p23.1), and R3 between markers D8S520 and D8S277 (8p23.1). All the tumors with partial losses showed deletions including one, two, or all three of these common regions of loss. Four cases (223, 486, 793, and 828) showed LOH only at marker D8S499, one case (984) only at D8S522, and one case (779) only at D8S1721. One case (1121) showed LOH at markers D8S552, D8S771, and D8S499 only and retained heterozygosity at marker D8S261 in between them. Several cases (484, 729, 733, 775, 818, 1105, 1106, and 1164) showing deletion of two or three of the minimal common regions of loss had markers between the regions that retained heterozygosity. Markers D8S261 and D8S520 showed the highest rate of retention of both alleles, and they were localized between the minimal common regions R1 and R2, and between R2 and R3, respectively. Figure 1B. shows an example of LOH results in a tumor where a part of chromosome 8p had been lost (Case 1121).
A, Deletion map of 20 serous ovarian carcinomas showing partial deletions (30%) at chromosome arm 8p. The 21 serous carcinomas with loss of heterozygosity (LOH) at all informative markers (34%) are not shown. The genetic order and the approximate loci of 18 microsatellite markers are shown on the right side of the chromosome 8p figure. Each vertical line represents one tumor sample; case numbers are shown at the top of each vertical line. Frequency of allelic loss (LOH/informative) at each marker is presented on the right. •, LOH; ○, informative with no loss; [50 percent shaded circle], not interpretable; —, not informative. Shaded area, potential deletions which include markers showing LOH and flanking noninformative markers. R1, R2, and R3, minimal common regions of loss. B, Representative example of LOH assessment; Case 1121: upper figure, marker D8S1107, informative with no loss; middle figure, marker D8S640, not informative (homozygous); lower figure, marker D8S552, LOH. Size in bp is shown on the X axis at the top of each figure. The peak heights in fluorescence units are shown on the Y axis on the right of each figure. In each figure the upper trace is amplification from normal tissue and the lower trace amplification from tumor tissue. Each peak has two labels (in boxes): upper label, size in bp; lower label, peak area.
Clinicopathological Characteristics and LOH at 8p
In serous carcinomas, LOH at 8p was detected in 36% of Grade 1 tumors, in 68% of Grade 2 tumors, and in 77% of Grade 3 tumors (p = 0.029). There was no correlation between LOH and stage of the serous tumor. In mucinous carcinomas, these clinicopathological parameters did not show an association with LOH.
Northern Blot Analysis of GATA-4
To characterize the expression of GATA-4 in mucinous and serous ovarian tissue samples, Northern analysis was performed. GATA-4 mRNA expression was detected in 16 of 26 (62%) mucinous and in 4 of 33 (12%) serous carcinoma specimens studied (p < 0.0001). Examples of GATA-4 mRNA expression in mucinous and serous carcinomas are shown in Figure 2.
An example of GATA-4 mRNA expression in mucinous (A) and serous (B) ovarian carcinoma specimens (n = 12). Fourteen micrograms total RNA from different tumor samples were subjected to Northern hybridization analysis with 32P-labeled GATA-4 cDNA probe. Glyceraldehyde-6-phosphate dehydrogenase (GAPDH) transcripts are shown as a control for even loading.
Immunohistochemistry of GATA-4 by Tissue Microarray
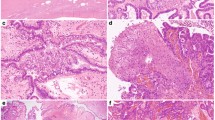
To evaluate expression of GATA-4 at the protein level immunohistochemical staining of GATA-4 was performed on ovarian carcinoma tissue microarray containing 75 mucinous and 528 serous ovarian carcinomas and 20 normal ovarian samples. Examples of GATA-4 immunostaining of tissue microarray are shown in Figure 3. In the normal ovary, GATA-4 was localized in the nuclei of surface epithelial cells. Particularly the cuboidal and columnar cells on the surface and in the inclusion glands were strongly immunoreactive. Positive nuclear staining of GATA-4 in carcinoma cells was present in 66% (49/75) of mucinous carcinomas and half of these cases showed moderate to strong immunostaining. However, most of the serous carcinomas had lost GATA-4 expression. Only 2.3% (12/528) of serous tumors showed positive immunostaining of carcinoma cells. The difference in the frequency of GATA-4 protein expression between mucinous and serous ovarian carcinomas was statistically highly significant (p < 0.0001) and it remained significant when only low grade (1–2; p < 0.0001) or low stage (I–II; p < 0.0001) tumors were taken into account. Positive immunostaining was noted in the nuclei of stromal cells in some of the samples (normal ovary, serous, and mucinous carcinomas), which is in accordance with our earlier results in the murine and human ovary (Heikinheimo et al, 1997; Laitinen et al, 2000).
Hematoxylin-eosin and immunohistochemical staining of GATA-4 on normal ovary and serous and mucinous carcinoma specimens on tissue microarray (original magnification, × 20 and × 100 in the inserts). A to C, Hematoxylin-eosin staining of normal ovarian tissue, mucinous carcinoma, and serous carcinoma, respectively. Immunoperoxidase staining shows GATA-4 protein in the nuclei of normal ovarian surface epithelial cells (D) and in those of mucinous carcinoma cells (E). Scant positive immunostaining of stromal cells is seen both in the normal ovary and mucinous carcinoma. In serous carcinoma, GATA-4 is absent from the tumor cells (F), and positive staining is found only in the stromal cells.
In mucinous carcinomas, GATA-4 staining correlated negatively with the grade and the stage of the tumors, ie, higher grade and higher stage tumors showed more often negative GATA-4 staining (p = 0.016 for grade, p < 0.0001 for stage). In serous carcinomas, there was no association between GATA-4 staining and grade or stage of the tumors. Forty-one of the 61 serous and 10 of the 14 mucinous tumors analyzed for LOH at distal 8p were included in the tumor microarray (Table 1). All the serous cases showing allelic loss of the whole distal 8p had lost GATA-4 protein expression, and only 1 of 7 cases showing partial deletions including GATA-4 region presented with positive GATA-4 staining. Of the 19 serous cases showing no LOH at distal 8p, only 2 were positive for GATA-4 staining. In mucinous carcinomas 2 cases showed allelic loss at one marker located at R3 (249 at D8S1140, 783 at D8S1721), and both of these cases were positive for GATA-4 immunostaining. The 7 of the 8 mucinous carcinomas showing no LOH at distal 8p had positive GATA-4 staining and, in one, staining was not interpretable. Different sample material was used for Northern blot analysis so comparison of mRNA expression data with LOH and protein expression results in each sample was not possible.
Discussion
The present results show that serous and mucinous ovarian carcinomas clearly differ with respect to the frequency and pattern of LOH at the distal part of the short arm of chromosome 8. In serous carcinoma, LOH was detected in 67% of the samples, whereas only 21% of mucinous carcinomas showed allelic loss. In serous carcinoma the typical aberration was loss of all or most of the informative markers, indicating loss of the entire distal 8p or large parts of it. In mucinous carcinomas, only one or two loci were affected in the three samples showing LOH, and the deleted markers were different in each sample, suggesting that they could be incidental. In many of the previous LOH studies on ovarian carcinoma, no differences have been detected between the histological types (Brown et al, 1999; Cliby et al, 1993; Wright et al, 1998; Yang-Feng et al, 1993), probably because only a few mucinous tumors have been included in these studies. However, in one allelotype study concerning all autosomal chromosome arms, there was a higher overall frequency of LOH in serous than in mucinous carcinomas (Sato et al, 1991), and LOH at 17p has been found to be typical of serous carcinoma but rare in mucinous carcinoma (Papp et al, 1996; Pierretti et al, 1995). The present results are in agreement with the increasing molecular genetic (Enomoto et al, 1991; Milner et al, 1993; Obata et al, 1998), subchromosomal (Tapper et al, 1997), and chromosomal (Diebold et al, 1997) evidence of distinct pathways of carcinogenesis in different histological types, notably serous and mucinous, of ovarian carcinoma.
Loss of heterozygosity at 8p has been considered to be an early event in carcinogenesis, as suggested by allelic losses detected in prostate intra-epithelial neoplasia, in situ ductal carcinoma of the breast, and preneoplastic bronchial epithelium (Anbazhagan et al, 1998; Emmert-Buck et al, 1995; Wistuba et al, 1999). However, LOH at 8p has also been found to associate with advanced stage tumors in ovarian and breast carcinomas (Brown et al, 1999; Wright et al 1998; Yokota et al, 1999). We found an association of LOH at 8p with tumor grade, but not with tumor stage. The diversity of the information obtained on allelic loss at 8p and clinicopathological parameters in ovarian and other tumor types suggests the presence of several tumor suppressor genes at this chromosomal arm involved in different phases of tumor development.
The frequency of LOH we observed (67%) is higher than previously reported at 8p (26–58%) and among the highest observed at any of the chromosomal arms in ovarian carcinoma (Brown et al, 1999; Cliby et al, 1993; Sato et al, 1991; Wright et al, 1998; Yang-Feng et al, 1993). However, previous studies have combined different histological types of ovarian carcinomas, whereas our result is based on serous tumors only. Previously, microsatellite instability (MSI) has been reported at a frequency of 50% in endometrioid ovarian carcinoma, but only 8% of serous tumors have shown MSI phenotype (Fujita et al, 1995). We found that only one serous tumor out of 62 exhibited instability of several microsatellites, thus meeting the criteria of an MSI phenotype (Boland et al, 1998). This suggests that mismatch-repair-system deficiency, whether inherited or sporadic, does not have a significant role in the molecular pathogenesis of serous ovarian carcinoma.
Three distinct minimal common regions of deletion could be defined by constructing a deletion map of the 20 serous tumors presenting with partial losses at distal 8p: R1 between D8S1810 and D8S1771 (8p21.1), R2 between D8S1731 and D8S640 (8p22-p23.1), and R3 between D8S520 and D8S277 (8p23.1). The results of previous LOH studies on 8p in ovarian carcinoma have been somewhat conflicting. One study reported three regions of overlapping deletions (two in 8p23 and one in 8p22) with 16 microsatellite markers in 9 tumors (Wright et al, 1998), whereas in another study involving 40 epithelial ovarian tumors and 6 microsatellite markers at 8p12–p22, the highest frequency of allelic loss was found at D8S136 (8p21) (Brown et al, 1999). The distal and middle regions of overlap in the study by Wright et al (1998) correspond to the R3 and the R2 regions in our study, respectively, and the proximal region is between R1 and R2 found in our study. The marker with highest frequency of LOH (D8S136) in the work by Brown et al (1999) is located between R1 and R2 detected in our study. In the two earlier studies, various histological subtypes of ovarian carcinoma have been analyzed in combination, and this makes a direct comparison of the results difficult. In neither of these studies was there an association between LOH at 8p and histological subtype of the tumor, which is probably due to the small number of mucinous tumors included. In these studies altogether, three mucinous carcinomas were analyzed and none of them showed LOH at 8p, whereas 33 of the 53 (62%) serous carcinomas presented with allelic loss at this region (Brown et al, 1999; Wright et al, 1998). Taken together, the complex pattern of allelic loss found in the present study and the literature suggests the presence of more than one tumor suppressor gene at distal 8p in ovarian carcinoma and especially in its serous histological type.
High-density mapping studies have defined several distinct minimal common regions of loss at 8p in various carcinomas, indicating the presence of two or more tumor suppressor genes (Adélaide et al, 1998; Farrington et al, 1996). However, only a few or no mutations have been found in the candidate genes at this region and no consistent tumor suppressor gene has been established at 8p to date. The most proximal minimal common region of loss in our study (R1), located at 8p21, is included in the commonly deleted regions in prostate, colon, and breast carcinomas (Adélaide et al, 1998; Farrington et al, 1996; Vocke et al, 1996). Deletions affecting more telomeric parts of 8p have also been reported in several types of carcinoma, including breast and lung carcinomas (Anbazhagan et al, 1998; Wistuba et al, 1999), which correspond to the R2 and R3 regions at 8p22–23.1 in our study. Three candidate tumor suppressor genes, PRLTS (platelet-derived growth factor [PDGF]-receptor beta-like tumor suppressor), N33, and EXTL3, have been isolated at 8p12–p22, but they are located outside the R1 and R2 regions in the present report (Fujiwara et al, 1995; MacGrogan et al, 1996; Van Hul et al, 1998). Another putative tumor suppressor gene, DLC1 (deleted in liver cancer), located at 8p21.3–p22, showed no mutations in 33 ovarian carcinomas (Wilson et al, 2000). Consequently, the genes mentioned above are unlikely to be the primary suspects as regards the losses we detected.
CGH and allelic analyses serve to narrow the regions of interest in the genome, but the identification of relevant genes is based on mutation, expression, and functional studies. GATA4 gene was chosen for mRNA and protein expression studies because it fulfills many features of a tumor suppressor gene putatively involved in the genesis of ovarian carcinoma. GATA4 gene is located at one of the minimal common regions of loss detected in our study (R3 at 8p23.1) and was known to be expressed in the nontransformed surface epithelium of mouse ovary (Heikinheimo et al, 1997). It is a zinc finger transcription factor that recognizes the consensus GATA motif present in the promoter of many genes involved in the differentiation and proliferation of a variety of tissues including the ovary (Evans, 1997). We detected GATA-4 expression in the normal surface epithelium of the ovary and in over 60% of mucinous carcinomas. However, most of the serous carcinomas had lost GATA-4 expression: only 12% and 2.3% of serous tumors showed positive mRNA and protein expression, respectively. There are several studies of the role of GATA-4 in organ development and tissue differentiation, but little is known about its function in tumorigenesis. Amplification of 8p22–p23 containing GATA4 gene was noted in a subset of esophageal (13.6%) and gastric cardia (12.5%) adenocarcinomas (Lin et al, 2000), but on the other hand, GATA-4 expression was lost in 4 of 11 gastric carcinoma cell lines (Bai et al, 2000). The role of GATA-4 in the pathogenesis of ovarian and other carcinomas remains to be determined. Of interest is the finding that, according to transactivation studies, GATA-4 directly regulates the expression of Müllerian inhibiting substance (Viger et al, 1998) and inhibin-α (Ketola et al, 1999), both putative tumor suppressor genes belonging to the TGF-β superfamily.
Forty-one serous and 10 mucinous carcinomas were analyzed for both LOH and GATA-4 protein expression. All mucinous carcinomas showing no LOH at R3 (D8S520–D8S277) expressed GATA-4 protein (except one in which immunostaining was not interpretable). Two mucinous carcinomas showed LOH at one R3 marker each, but they still expressed GATA-4, which suggests that the deletions in these tumors may not include GATA4 locus or that the other allele is still active. Twenty-one of 22 serous carcinomas showing allelic loss at R3 had lost GATA-4 expression. Seventeen of 19 serous tumors showing no LOH at this region were also negative for GATA-4 expression. In these tumors small deletions may reside between the markers used in the study or the mechanism of down-regulated expression may not involve allelic loss. Inactivation of a tumor suppressor without allelic loss is seen for example in MSI-positive sporadic colorectal carcinoma, where hMLH1 is frequently inactivated by biallelic hypermethylation of the gene's promoter or by mutation of one allele and hypermethylation of the other allele (Wheeler et al, 1999). In the future, knowledge of the genomic sequence of the commonly lost regions will enable mutation and other analyses of GATA4 and other genes at distal 8p in serous ovarian carcinoma.
In conclusion, we found frequent allelic loss at 8p21–p23 in serous but not in mucinous ovarian carcinomas. In serous carcinoma, three distinct minimal common regions of loss could be defined. As a rule, expression of the transcription factor gene GATA4, located at one of these regions (8p23.1), was found to be lost in serous but retained in mucinous carcinomas. However, the complex pattern of allelic loss suggests the presence of more than one tumor suppressor gene at distal 8p in serous ovarian carcinoma. Further research is needed to characterize the role of GATA4 and to identify other putative suppressor genes located at this region in serous ovarian carcinoma. Our finding that serous and mucinous ovarian carcinomas differ with respect to molecular changes at distal 8p indicates different mechanisms of tumorigenesis in these histological types of ovarian carcinoma. Appropriate categorization of ovarian carcinoma into biologically meaningful entities is a prerequisite for successful development of novel therapeutic and other intervention modalities in the future.
Materials and Methods
LOH
Tumor Samples, Microdissection, and DNA Extraction.
Tumor and blood samples were taken from 76 patients undergoing primary surgery for ovarian carcinoma at the Department of Obstetrics and Gynecology, Helsinki University Central Hospital. The tumors were staged according to the classification scheme of the International Federation of Gynecologists and Obstetricians (FIGO). All the specimens were reviewed by the same investigator (RB) as regards histological subtype and grade. There were 14 tumor samples of mucinous and 62 samples of serous histology. Of the mucinous carcinomas, 13 were Stage I and one was Stage II. Seven mucinous tumors were Grade 1, five were Grade 2, and two were Grade 3. Of the serous carcinomas, 14 were Stage I, 5 were Stage II, 38 were Stage III, and 5 were Stage IV. Twelve serous tumors were Grade 1, 19 were Grade 2, and 31 were Grade 3.
After removal, the tissues were snap-frozen. The proportion of carcinoma cells in the samples was evaluated by microscopy, and only tissue samples estimated to contain more than 40% to 50% of tumor cells were included in the study. In mucinous carcinomas, as a rule, the amount of nonneoplastic cells was high, and a laser microbeam microdissection technique (Schütze and Lahr, 1998) was used to increase the “purity” of the samples before DNA extraction. In the serous carcinomas the amount of contaminating stromal cells was low, and no microdissection was needed for these samples. Tumor DNA and normal DNA were extracted from fresh-frozen tumor samples and from blood lymphocytes, respectively, according to a proteinase K-phenol-chloroform method.
Microsatellites.
A set of 18 highly polymorphic microsatellite markers was used to determine LOH status at the distal part of the short arm of chromosome 8 (8p21–p23). Primer sequences and reaction conditions for dinucleotide markers were obtained from the Genethon (http://ftp.genethon.fr) human linkage map (D8S258, D8S261, D8S262, D8S277, D8S283, D8S520, D8S552, D8S560, D8S1721, D8S1810, D8S1824, D8S1731, D8S1771, and D8S1781). Tetranucleotide markers came from the Genome Database (http://gdbwww.gdb.org) (D8S499, D8S640, D8S1107, and D8S1140). The genetic order of the markers was based on the Genethon map (Fig. 1A). According to GeneMap'99, the most probable location of GATA4 is between the markers D8S520 and D8S277 (distance 11.3 cM), which were included in the present study. The oligonucleotides were labeled fluorescently with one of three dyes (6-FAM, TET, HEX; Institute of Biotechnology, University of Helsinki, Finland). A fourth dye (TAMRA; Perkin-Elmer, Foster City, California) was reserved for the size standard.
PCR.
The PCR reactions for fluorescent markers were carried out in a volume of 10 μl and included GeneAmp 1 × PCR buffer (Perkin-Elmer), each dNTP at 50 μmol/l, 60 ng DNA, 0.5 U AmpliTaq Gold polymerase (Perkin-Elmer), and 5 pmol of each primer (one of them fluorescently labeled). Of the microdissected samples, 5 to 10 ng of DNA was used for the PCR reaction. The reaction mixtures were given 30 to 35 cycles of 5 seconds at 96° C, 59 seconds at 92° C, 1 minute 15 seconds at 55° C, and 45 seconds at 72° C, preceded by a 10-minute hot start at 96° C for enzyme activation and followed by final extension at 72° C for 30 minutes.
Electrophoresis and Allele Scoring.
The 18 fluorescent products for each template were pooled in two groups, each consisting of 9 μl. Overlapping size ranges of products with the same label could be avoided by combining loci D8S258, D8S262, D8S277, D8S522, D8S560, D8S1107, D8S1781, D8S1810, and D8S1824 in one group, and loci D8S261, D8S283, D8S499, D8S520, D8S640, D8S1140, D8S1721, D8S1731, and D8S1771 in the other group. One μl of this mixture was added to 12.5 μl formamide and 0.5 μl TAMRA 500 size standard and denatured at 96° C for 3 minutes before loading the samples into an ABI Prism 310 Genetic Analyzer (Perkin-Elmer), which introduces the samples into a polymer-filled capillary for electrophoresis. As the labeled samples travel through the capillary, they are excited by the laser and the emitted light is detected and analyzed by GeneScan software (Perkin-Elmer). Finally, Genotyper software (Perkin-Elmer) was used for allele scoring and assessment of LOH. Each fluorescent peak was quantified in terms of size (base pairs), peak height, and peak area. The peaks of the normal DNA sample were used to determine whether the sample was homozygous (one peak only) or heterozygous (two peaks). The sizes of the allele peaks were assigned according to the area under the highest peak. When two alleles were present in normal tissue and one was absent in the tumor, the result was determined to be LOH and no further calculation was performed. In cases where the assessment was not clear-cut, the ratio of alleles was calculated for each normal and tumor sample, and the tumor ratio was divided by the normal ratio, ie, T2:T1/N2:N1 (T1 and N1 are the area values for the shorter length alleles, and T2 and N2 are the values for the longer length alleles, for tumor and normal tissue respectively). If the ratio was less than 0.6 or greater than 1.67, then one of the alleles had decreased more than 40% and the result was determined as LOH (Canzian et al, 1996). In ambiguous cases, the PCR, electrophoresis, and scoring were repeated.
GATA-4 Northern Analysis
RNA from ovarian carcinoma samples was extracted and Northern blotting was performed as previously described (Laitinen et al, 1997). As probes for filter hybridizations, we used a human GATA-4 cDNA (White et al, 1995) and a rat glyceraldehyde-6-phosphate dehydrogenase (GAPDH) cDNA (Laitinen et al, 1997). The cDNAs were labeled with [32P]-α-deoxy-CTP and a Prime-a-gene kit (Promega, Madison, Wisconsin). Expression of GATA-4 mRNA was studied in 33 serous and 26 mucinous ovarian carcinoma specimens.
Ovarian Carcinoma Tissue Microarray
Formalin-fixed and paraffin-embedded tumor specimens from years 1964 to 1999 and control normal ovarian surface epithelium were obtained from the archives of the Department of Obstetrics and Gynecology, Helsinki University Central Hospital. The specimens included 528 serous and 75 mucinous primary ovarian carcinomas and 20 normal ovarian samples. All tumors and controls were reviewed by one pathologist (RB). The grade and stage distribution of serous carcinomas was as follows: Grade 1 (41%), Grade 2 (23%), Grade 3 (36%), and Stage I (26%), Stage II (11%), Stage III (50%), and Stage IV (13%). The distribution in mucinous carcinomas was as follows: Grade 1 (71%), Grade 2 (25%), Grade 3 (4%), and Stage I (77%), Stage II (4%), Stage III (17%), and Stage IV (1.3%).
The tissue microarrays were constructed as described previously (Kononen et al, 1996). In brief, a representative tumor area was selected from hematoxylin-eosin-stained sections of each tumor. Core tissue biopsy specimens (diameter 0.8 mm) were taken from these areas of individual donor blocks and precisely arrayed into a new recipient paraffin block with a custom-built precision instrument (Beecher Instruments, Silver Spring, Maryland). Four core tissue biopsies were obtained from each carcinoma specimen. After the block construction was completed, 5-μm sections were cut with a microtome. The presence of tumor tissue on the arrayed samples was verified on hematoxylin-eosin-stained section.
GATA-4 Immunohistochemistry
The sections of tissue microarray were fixed in 4% paraformaldehyde and subjected to immunohistochemistry using a goat polyclonal anti-mouse GATA-4 IgG (Santa Cruz Biotechnology, Santa Cruz, California) as the primary antibody and a nonimmune goat IgG as control (Heikinheimo et al, 1997). This antibody recognizes human as well as mouse GATA-4. A commercially available avidin-biotin immunoperoxidase system was used to visualize bound antibody (Vectastain Elite ABC Kit; Vector Laboratories, Burlingame, California). 3-Amino-9-ethylcarbazole (Sigma Chemicals, St. Louis, Missouri) was used as the chromogen, and the development reaction took place in the presence of 0.03% H2O2. The sections were counterstained with 10% hematoxylin. Clear nuclear staining was required for the result to be interpreted as positive.
Statistical Analysis
Statistical analyses were performed by using Fisher's exact test (two-tailed p-values) and the nonparametric Mann-Whitney U test.
References
Adélaide J, Chaffanet M, Imbert A, Allione F, Geneix J, Popovici C, van Alewijk D, Trapman J, Zeillinger R, Børresen-Dale A-L, Lidereau R, Birnbaum D, and Pébusque M-J (1998). Chromosome region 8p11-p21: Refined mapping and molecular alterations in breast cancer. Genes Chromosomes Cancer 22: 186–199.
Anbazhagan R, Fujii H, and Gabrielson E (1998). Allelic loss of chromosomal arm 8p in breast cancer progression. Am J Pathol 152: 815–819.
Bai Y-Q, Akiyama Y, Nagasaki H, Yagi OK, Kikuchi Y, Saito N, Takeshita K, Iwai T, and Yuasa Y (2000). Distinct expression of CDX2 and GATA4/5, development-related genes, in human gastric cancer cell lines. Mol Carcinog 28: 184–188.
Boland RC, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, and Srivastava SA (1998). National Cancer Institute Workshop on microsatellite instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58: 5248–5257.
Brown MR, Chuaqui R, Vocke CD, Berchuck A, Middletown LP, Emmert-Buck MR, and Kohn EC (1999). Allelic loss on chromosome arm 8p: Analysis of sporadic epithelial ovarian tumors. Gynecol Oncol 74: 98–102.
Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick RB, Aaltonen LA, and de la Chapelle A (1996) Semiautomated assessment of loss of heterozygosity and replication error in tumors. Cancer Res 56: 3331–3337.
Cliby W, Ritland S, Hartmann L, Dodson M, Halling KC, Keeney G, Podratz KC, and Jenkins RB (1993). Human epithelial ovarian cancer allelotype. Cancer Res 53: 2393–2398.
Diebold J, Siegert S, Baretton GB, Suchy B, Meier W, Haas CJ, and Lohrs U (1997). Interphase cytogenetic analysis of mucinous ovarian neoplasms. Lab Invest 76: 661–670.
Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Liotta LA, and Linehan WM (1995). Allelic loss on chromosome 8p12–21 in microdissected prostatic intraepithelial neoplasia. Cancer Res 55: 2959–2962.
Enomoto T, Weghorst CM, Inoue M, Tanizawa O, and Rice JM (1991). K-ras activation occurs frequently in mucinous adenocarcinomas and rarely in other common epithelial tumors of the human ovary. Am J Pathol 139: 777–785.
Evans T (1997). Regulation of cardiac gene expression by GATA-4/5/6. Trends Card Med 7: 75–83.
Farrington SM, Cunningham C, Boyle SM, Wyllie AH, and Dunlop MG (1996). Detailed physical and deletion mapping of 8p with isolation of YAC clones from tumour suppressor loci involved in colorectal cancer. Oncogene 12: 1803–1808.
Fujita M, Enomoto T, Yoshino K, Nomura T, Buzard GS, Inoue M, and Okudaira Y (1995). Microsatellite instability and alterations in the hMSH2 gene in human ovarian cancer. Int J Cancer 64: 361–366.
Fujiwara Y, Ohata H, Kuroki T, Koyama K, Tsuchiya E, Monden M, and Nakamura Y (1995). Isolation of a candidate tumor suppressor gene on chromosome 8p21.3-p22 that is homologous to an extracellular domain of the PDGF receptor beta gene. Oncogene 10: 891–895.
Heikinheimo M, Ermolaeva M, Bielinska M, Rahman NA, Narita N, Huhtaniemi IT, Tapanainen JS, and Wilson DB (1997). Expression and hormonal regulation of transcription factors GATA-4 and GATA-6 in the mouse ovary. Endocrinology 138: 3505–3514.
Jenkins RB, Bartelt D Jr, Stalboerger P, Persons D, Dahl RJ, Podratz K, Keeney G, and Hartmann L (1993). Cytogenetic studies of epithelial ovarian carcinoma. Cancer Genet Cytogenet 71: 76–86.
Ketola I, Rahman N, Toppari J, Bielinska M, Porter-Tinge SB, Tapanainen JS, Huhtaniemi IT, Wilson DB, and Heikinheimo M (1999). Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 140: 1470–1478.
Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, and Kallioniemi OP (1996). Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 4: 844–847.
Laitinen M, Ristimäki A, Honkasalo M, Narko K, Paavonen K, and Ritvos O (1997). Differential hormonal regulation of vascular endothelial growth factors VEGF, VEGF-B, and VEGF-C messenger ribonucleic acid levels in cultured human granulosa-luteal cells. Endocrinology 138: 4748–4756.
Laitinen MP, Anttonen M, Ketola I, Wilson DB, Ritvos O, Butzow R, and Heikinheimo M (2000). Transcription factors GATA-4, GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord derived ovarian tumors. J Clin Endocrinol Metab 85: 3476–3483.
Lin L, Aggarwal S, Glover TW, Orringer MB, Hanash S, and Beer DG (2000). A minimal critical region of the 8p22–23 amplicon in esophageal adenocarcinomas defined using sequence tagged site- amplification mapping and quantitative polymerase chain reaction includes the GATA-4 gene. Cancer Res 60: 1341–1347.
MacGrogan D, Levy A, Bova GS, Isaacs WB, and Bookstein R (1996). Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics 35: 55–65.
Makar AP, Baekelandt M, Trope CG, and Kristensen GB (1995). The prognostic significance of residual disease, FIGO substage, tumor histology, and grade in patients with FIGO stage III ovarian cancer. Gynecol Oncol 56: 175–180.
Milner BJ, Allan LA, Eccles DM, Kitchener HC, Leonard RCF, Kelly KF, Parkin DE, and Haites NE (1993). p53 mutation is a common genetic event in ovarian carcinoma. Cancer Res 53: 2128–2132.
Obata K, Morland SJ, Watson RH, Hitchcock A, Chenevix-Trench G, Thomas EJ, and Campbell IG (1998). Frequent PTEN/MMAC mutations in endometrioid but not serous or mucinous epithelial ovarian tumors. Cancer Res 58: 2095–2097.
Omura GA, Brady MF, Homesley HD, Yordan E, Major FJ, Buchsbaum HJ, and Park RC (1991). Long-term follow-up and prognostic factor analysis in advanced ovarian carcinoma: The Gynecologic Oncology Group experience. J Clin Oncol 9: 1138–1150.
Papp J, Csokay B, Bosze P, Zalay Z, Toth J, Ponder B, and Olah E (1996). Allele loss from large regions of chromosome 17 is common only in certain histological subtypes of ovarian carcinomas. Br J Cancer 74: 1592–1597.
Pejovic T, Heim S, Mandahl N, Baldetorp B, Elmfors B, Flodérus U-M, Furgyik S, Helm G, Himmelmann A, Willén H, and Mitelman F (1992). Chromosome aberrations in 35 primary ovarian carcinomas. Genes Chromosomes Cancer 4: 58–68.
Pere H, Tapper J, Seppälä M, Knuutila S, and Butzow R (1998). Genomic alterations in fallopian tube carcinoma: Comparison to serous uterine and ovarian carcinomas reveals similarity suggesting likeness in molecular pathogenesis. Cancer Res 58: 4274–4276.
Pierretti M, Cavalieri C, Conway PS, Gallion HH, Powell DE, and Turker MS (1995). Genetic alterations distinguish different types of ovarian tumors. Int J Cancer 64: 434–440.
Risch HA, Marret LD, Jain M, and Howe GR (1996). Differences in risk factors for epithelial ovarian cancer by histologic type. Results of a case-control study. Am J Epidemiol 144: 363–372.
Sato T, Saito H, Morita R, Koi S, Lee JH, and Nakamura Y (1991). Allelotype of human ovarian cancer. Cancer Res 51: 5118–5122.
Schütze K and Lahr G (1998). Identification of expressed genes by laser-mediated manipulation of single cells. Nat Biotechnol 16: 737–742.
Tapper J, Butzow R, Wahlström T, Seppälä M, and Knuutila S (1997). Evidence for divergence of DNA copy number changes in serous, mucinous and endometrioid ovarian carcinomas. Br J Cancer 75: 1782–1787.
Van Hul W, Wuyts W, Hendrickx J, Speleman F, Wauters J, De Boulle K, Van Roy N, Bossuyt P, and Willems PJ (1998). Identification of a third EXT-like gene (EXTL3) belonging to the EXT gene family. Genomics 47: 230–237.
Viger RS, Mertineit C, Trasler JM, and Nemer M (1998). Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Müllerian inhibiting substance promoter. Development 125: 2665–2675.
Vocke CD, Pozzati RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Liotta LA, Emmert-Buck MR, and Linehan WM (1996). Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12–21. Cancer Res 56: 2411–2416.
Wheeler JMD, Beck NE, Kim HC, Tomlinson IPM, Mortensen NJM, and Bodmer WF (1999). Mechanisms of inactivation of mismatch repair genes in human colorectal cancer cell lines: The predominant role of hMLH1. Proc Natl Acad Sci USA 96: 10296–10301.
White RA, Dowler LL, Pasztor LM, Gatson LL, Adkison LR, Angeloni SV, and Wilson DB (1995). Assignment of the transcription factor GATA4 gene to human chromosome 8 and mouse chromosome 14: Gata4 is a candidate gene for Ds (disorganization). Genomics 27: 20–26.
Wilson PJ, McGlinn E, Marsh A, Evans T, Arnold J, Wright K, Biden K, Young J, Wainwright B, Wicking C, and Chenevix-Trench G (2000). Sequence variants of DLC1 in colorectal and ovarian tumours. Hum Mutat 15: 156–165.
Wistuba II, Behrens C, Virmani AK, Milchgrub S, Syed S, Lam S, Mackay B, Minna JD, and Gazdar AF (1999). Allelic losses at chromosome 8p21–23 are early and frequent events in the pathogenesis of lung cancer. Cancer Res 59: 1973–1979.
Wright K, Wilson PJ, Kerr J, Do K, Hurst T, Khoo S-K, Ward B, and Chenevix-Trench G (1998). Frequent loss of heterozygosity and three critical regions on the short arm of chromosome 8 in ovarian adenocarcinomas. Oncogene 17: 1185–1188.
Yang-Feng TL, Han H, Chen K-C, Li S, Claus EB, Carcangiu ML, Chambers SK, Chambers JT, and Schwartz PE (1993). Allelic loss in ovarian cancer. Int J Cancer 54: 546–551.
Yokota T, Yoshimoto M, Akiyama F, Sakamoto G, Kasumi F, Nakamura Y, and Emi M (1999). Localization of a tumor suppressor gene associated with the progression of human breast carcinoma within a 1-cM interval of 8p22-p23.1. Cancer 85: 447–452.
Acknowledgements
This work was supported by grants from the Helsinki University Central Hospital Research Fund, Finnish Medical Foundation, Finnish Cancer Society, Sigrid Juselius Foundation, Foundation of Jalmari and Rauha Ahokas, Academy of Finland, and University of Helsinki Research Funds.
Ms. Gynel Arifdshan, Ms. Ritva Javanainen, Ms. Marjo Rissanen, and Ms. Anita Saarinen are warmly thanked for excellent technical assistance. Dr. Patrik Lassus is thanked for his help in statistics. Dr. David B. Wilson is acknowledged for his critical review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lassus, H., Laitinen, M., Anttonen, M. et al. Comparison of Serous and Mucinous Ovarian Carcinomas: Distinct Pattern of Allelic Loss at Distal 8p and Expression of Transcription Factor GATA-4. Lab Invest 81, 517–526 (2001). https://doi.org/10.1038/labinvest.3780260
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780260
This article is cited by
-
Effect of Helicobacter pylori Infection on GATA-5 and TFF1 Regulation, Comparison Between Pediatric and Adult Patients
Digestive Diseases and Sciences (2018)
-
Genetic Alterations of Chromosome 8 Genes in Oral Cancer
Scientific Reports (2014)
-
The overexpression of MCPH1 inhibits cell growth through regulating cell cycle-related proteins and activating cytochrome c-caspase 3 signaling in cervical cancer
Molecular and Cellular Biochemistry (2014)
-
Methylation Pattern of THBS1, GATA-4, and HIC1 in Pediatric and Adult Patients Infected with Helicobacter pylori
Digestive Diseases and Sciences (2013)
-
GATA Factors in Gastrointestinal Malignancy
World Journal of Surgery (2011)