Abstract
TNF signals are mediated through two different receptors, TNFR1 and TNFR2. In endothelial cells, TNFR1 is predominantly localized in the Golgi apparatus and TNFR2 on the plasma membrane. To investigate structural features responsible for the disparate localization, endothelial cells were transfected with epitope-tagged or green fluorescent protein-fused wild type and mutant receptor molecules. Wild type receptors recapitulated the distribution of endogenous receptors. Deletions of the entire TNFR1 intracellular domain or of the C-terminal death domain (TNFR1−DD) allowed expression of the receptor on the plasma membrane. However, addition of the death domain to the C-terminus of TNFR2 (TNFR2+DD) did not lead to Golgi-retention of this chimeric receptor. Overexpressed TNFR1, TNFR2, and TNFR2+DD increased basal expression of a cotransfected NF-κB–dependent promotor-reporter gene. Overexpressed TNFR1−DD did not activate NF-κB but acted as a ligand-specific dominant negative inhibitor of TNF actions. Unexpectedly, TNF responses were also inhibited by overexpressed TNFR1 and TNFR2+DD, but not TNFR2. We conclude that the death domain of TNFR1 is required for retention of TNFR1 in the Golgi apparatus but is not sufficient to direct Golgi retention of a TNFR2+DD chimera, and that overexpressed receptors that contain the death domain (TNFR1 and TNFR2+DD) spontaneously activate NF-κB while inhibiting TNF responses.
Similar content being viewed by others
Introduction
Tumor necrosis factor (TNF) is a major mediator of inflammatory responses. It is a homotrimer produced mainly by activated macrophages and T lymphocytes. TNF binds to two single chain receptors, TNF receptor 1 (TNFR1, CD120a) or TNF receptor 2 (TNFR2, CD120b), both of which are expressed on most cell types. Binding of TNF to monomeric receptors on the plasma membrane of target cells induces homotypic receptor trimerization [ie, formation of (TNFR1)3 or of (TNFR2)3]. The intracellular domains of these clustered receptor monomers bind intracellular adaptor molecules, which in turn initiate signaling cascades leading either to cell activation or cell death (reviewed in Tartaglia and Goeddel, 1992b). Receptors mutated to remove the intracellular regions may act as dominant negative mutants, presumably by forming receptor complexes that are unable to recruit the relevant adaptor molecules (Brakebusch et al, 1992; Tartaglia and Goeddel, 1992a).
TNFR1 and TNFR2 both belong to the large TNFR superfamily defined by homologies in the extracellular domains of its members, which include Fas, CD40, and nerve growth factor receptor, among others (Bazzoni and Beutler, 1996; Smith et al, 1994). In contrast to their extracellular domains, the intracellular domains of TNFR1 and TNFR2 are not homologous and interact with different sets of adaptor molecules. The C-terminal half of the intracellular region of TNFR1 contains a protein-protein interaction motif dubbed the “death domain” that is homologous with other proteins involved in induction of apoptosis (Cleveland and Ihle, 1995; Feinstein et al, 1995; Tartaglia et al, 1993). Surprisingly, this region of TNFR1 has been shown to be necessary for TNF-mediated activation of gene transcription as well as death responses (Adam et al, 1996; Brakebusch et al, 1992; Machleidt et al, 1996; Tartaglia et al, 1993). The death domain of TNFR1 interacts with the death domain of the adaptor protein TRADD (TNF receptor-associated death domain protein), which is the first step in signal transduction leading both to the activation of the transcription factors NF-κB and AP-1 and to the activation of apoptosis-mediating caspases (Hsu et al, 1995; Park and Baichwal, 1996). Initiation of gene transcription is dependent upon recruitment of two additional adaptor proteins, RIP (receptor-interacting protein) and TRAF-2 (TNF receptor-associated factor 2) (Hsu et al, 1996a), each of which binds to TRADD and to each other. The death pathway is instead dependent upon interactions of TRADD with the adaptor protein FADD (Fas-associated death domain protein) (Hsu et al, 1996b). TNFR2 does not contain a death domain and instead interacts directly with the adaptor protein TRAF-2 via a degenerate peptide motif (SXXE) in its intracellular region (Park et al, 1999). In some cell types, recruitment of TRAF-2 by TNFR2 seems sufficient to cause the activation of NF-κB and AP-1 (Haridas et al, 1998); it is not known whether this response also involves RIP.
The vascular endothelium, which forms a barrier separating blood from the underlying tissues, is a principal target for TNF actions. Specifically, endothelial cells (EC) are activated by TNF binding to produce chemokines, such as MCP-1 (Strieter et al, 1989) and IL-8 (Strieter et al, 1988), and to express the adhesion molecules E-selectin (Bevilacqua et al, 1987; Pober et al, 1986), ICAM-1 (Pober et al, 1987), and VCAM-1 (Carlos et al, 1990; Osborn et al, 1989; Rice and Bevilacqua, 1989). These endothelial responses lead to the recruitment and stimulation of leukocytes, resulting in local inflammation with the possibility of tissue injury (Pober and Cotran, 1990).
TNF activation of EC is mostly dependent on TNFR1, although there is evidence that TNFR2 may participate in signaling at suboptimal TNF concentrations, perhaps through ligand passing to TNFR1 (Mackay et al, 1993; Slowik et al, 1993; Tartaglia et al, 1993). Of relevance to the “ligand passing” hypothesis, the two receptors reside largely in different cellular compartments, with TNFR1 predominantly localized to the Golgi apparatus and minimally expressed on the cell surface, and TNFR2 almost exclusively localized to the plasma membrane (Bradley et al, 1995). The significance of the intracellular sequestration of TNFR1 is not clear. Fas, another death domain-containing member of the TNFR superfamily, has been shown to be associated with Golgi in vascular smooth muscle cells; it is thought that this compartmentalization may be a way for cells to down-modulate ligand sensitivity (Bennett et al, 1998).
The present study was undertaken to investigate what structural features could be responsible for the disparate localization of the TNF receptors. Our strategy was to overexpress various epitope-tagged receptor constructs in human umbilical vein endothelial cells (HUVEC) and use both fluorescence-activated cell sorter (FACS) analysis and confocal fluorescence microscopy to localize the positions of the exogenous receptor molecules. We also evaluated the consequences of transfected receptor overexpression for basal and TNF-induced transcriptional responses with a cotransfected NF-κB–sensitive promotor-reporter gene. We show that the death domain of TNFR1, necessary for signaling through this receptor, is also necessary, but not sufficient, for Golgi retention. Furthermore, we show that overexpression above endogenous levels of TNFR1, but not of TNFR2, has an inhibitory effect on TNF signaling in EC, and that part of this inhibitory effect is mediated through the death domain.
Results
Transfected TNFR1 and TNFR2 in HUVEC Recapitulate the Distribution Pattern of the Endogenous TNF Receptors
By confocal fluorescence microscopy, endogenous TNFR1 and TNFR2 in EC have disparate intracellular localizations, with TNFR1 found mostly in the Golgi apparatus and TNFR2 found almost exclusively on the plasma membrane (Bradley et al, 1995; Gaeta ML, unpublished observations). To quantify this distribution, we used indirect immunofluorescence and FACS analysis of intact and permeabilized cells. As shown in Figure 1, 88% of TNFR1 can be detected only after permeabilization. In contrast, permeabilization only exposed an additional 17% of TNFR2. These numbers may be overestimated because a portion of the surface receptors may have been extracted during permeabilization. Nevertheless, these FACS data are consistent with the microscopy results, confirming that the majority of TNFR1 resides intracellularly, whereas most TNFR2 is on the cell surface.
Surface and intracellular expression of endogenous TNFR1 and TNFR2. HUVEC were harvested by collagenase treatment and examined by indirect immunofluorescence for TNFR1 (thick line), TNFR2 (dashed line), or irrelevant control mAb (K16/16, thin line) in nonpermeabilizing and permeabilizing conditions. Left panel, Cell surface expression of the TNF receptors in nonpermeabilized cells. Right panel, Combined intracellular and cell surface expression of the TNFR in permeabilized cells. The corrected mean fluorescence intensity (cMFI) was determined by subtraction of K16/16 and allows comparisons of fluorescence intensity between permeabilized and nonpermeabilized cells. Surface expression of TNFR1 reflects only a small fraction of the total pool, an additional 88% is detectable upon permeabilization, whereas 83% of TNFR2 is detectable without permeabilization.
To study the structural basis of these distribution patterns, we made expression constructs of the human TNF receptors with an epitope tag on the extracellular N-terminus and used these constructs to transiently transfect HUVEC. Green fluorescent protein (GFP) was cotransfected as an independent marker to identify the subset of EC (approximately 10% to 20%) that had been transfected. In our initial experiments, HUVEC were harvested 60 hours after transfection, immunoreacted with the anti-FLAG antibody using permeabilizing or nonpermeabilizing conditions, and analyzed by FACS. By restricting FACS analysis to those cells that expressed GFP, we quantified the number of cells displaying epitope-tagged receptors before and after permeabilization. As shown in Table 1, only 2.0% of intact cells transfected with TNFR1 were immunoreactive to the anti-FLAG antibody, whereas, after permeabilization, the fraction rose to 34.1%. In contrast, TNFR2-transfected cells were 55.8% immunoreactive before permeabilization and 62.2% after permeabilization. The observation that the number of immunoreactive permeabilized cells did not reach 100% is explained by the fact that the GFP label was much more readily detected than transfected receptors, and the GFP-immunoreactive population included numerous cells that expressed too few FLAG-tagged receptor molecules to be detected above background. Nevertheless, these data demonstrate that in HUVEC, transfected TNFR2 was readily expressed on the cell surface, whereas transfected TNFR1 was not, consistent with the results obtained from FACS analysis of the endogenous receptors.
To more precisely ascertain the localization pattern of the recombinant receptors, we examined the transfected cells by conventional and confocal immunofluorescence microscopy. HUVEC transfected in 100 mm plates were replated onto fibronectin-coated coverslips 48 hours after transfection. The cells were allowed to rest 12 hours, then fixed and immunoreacted with anti-FLAG antibody using permeabilizing or nonpermeabilizing conditions. We observed that FLAG-TNFR1 was essentially confined to the perinuclear Golgi region and not detectable on the cell surface. The Golgi localization of intracellular FLAG-TNFR1 was confirmed by co-localization with mannosidase II (data not shown). In contrast, FLAG-TNFR2 was highly expressed on the cell surface of unpermeabilized cells as well as in the perinuclear region of permeabilized cells (Fig. 2). The intracellular component of transfected TNFR2 molecules is likely to represent newly synthesized molecules. Thus, immunofluorescence microscopy confirmed that exogenously expressed TNFR molecules were localized in a similar pattern to the endogenous receptors.
Disparate localization patterns of transfected TNFR molecules in fixed HUVEC. HUVEC were transfected with full-length TNFR1 (FLAG-TNFR1, A and B), TNFR1 lacking the intracellular domain (FLAG-TNFR1−IC, C and D), TNFR1 lacking the death domain (FLAG-TNFR1−DD, E and F), or full-length TNFR2 (FLAG-TNFR2, G and H), fixed in 2% paraformaldehyde and examined by indirect immunofluorescence using an anti-FLAG mAb in nonpermeabilizing (A, C, E, and G) or permeabilizing (B, D, F, and H) conditions. Full-length TNFR1 is detectable in the perinuclear region but not on the cell surface, whereas the truncated TNFR1 constructs and full-length TNFR2 are detectable both on the surface and in the intracellular compartment. Photomicrographs are representative of cells from at least five different experiments.
Death Domain of TNFR1 Is Necessary but Not Sufficient for Golgi Retention
To determine whether the intracellular region of TNFR1, which differs from that of TNFR2, was responsible for retention of this receptor in the Golgi, we transfected HUVEC with a deletion construct, FLAG-TNFR1−IC, which consisted of the FLAG-tagged extracellular region and the transmembrane domain, but with the entire intracellular region truncated except for the first amino acid. In contrast to TNFR1, FLAG-TNFR1−IC was well distributed on the cell surface in nonpermeabilized cells. In permeabilized cells, the truncated receptor was also found in the perinuclear region (Fig. 2), presumably representing newly synthesized molecules.
To determine whether the death domain, which is located within the region responsible for localization of TNFR1 to the Golgi, is necessary for the regulation of the cellular distribution of TNFR1, we made a deletion construct, FLAG-TNFR1−DD, consisting of the FLAG-tagged extracellular domain, the transmembrane domain, and the membrane-proximal portion of the intracellular domain of TNFR1, truncating only the C-terminal half of the intracellular domain that comprises the death domain. In contrast to full-length TNFR1, TNFR1−DD was clearly detected on the cell surface (Fig. 2), indicating that the death domain was necessary for retention of TNFR1 in the Golgi.
Immunofluorescence results may be subject to artifacts introduced by the fixation and permeabilization necessary for antibody immunoreactivity. In particular, detergent solubilization may extract plasma membrane proteins. To circumvent this, we made GFP fusion constructs with GFP added at the C-terminus of TNFR1, TNFR2, and TNFR1−DD. GFP has the advantage of intrinsic fluorescence that permits its direct visualization using fluorescence microscopy, obviating the need for permeabilization and fixation and its possible perturbation of the cells. In this manner, the intracellular localization of gfpTNFR1, gfpTNFR2, and gfpTNFR1−DD were observed in living cells (Fig. 3, A to C). Whereas expression gfpTNFR1 was confined to the Golgi, both gfpTNFR1−DD and gfpTNFR2 were distributed to the plasma membrane and the intracellular compartment, including the Golgi and transport vesicles apparently trafficking to the surface. These results confirm the localization patterns detected by antibody immunoreactivity and FACS.
Disparate localization patterns of the green fluorescent protein (GFP) fusion TNFR molecules in live HUVEC. HUVEC transfected with GFP fusion TNFR constructs were examined live using a Zeiss LSM510 confocal microscope. Full-length TNFR1 (gfp-TNFR1, A) is confined to the perinuclear region and not detectable on the cell surface, whereas TNFR1 lacking the death domain (gfp-TNFR1−DD, B) is redistributed in the intracellular compartment and to the cell surface. Full-length TNFR2 (gfp-TNFR2, C) and the chimera TNFR2 with the death domain of TNFR1 (gfp-TNFR2+DD, D) show similar intracytoplasmic and surface expression. Photomicrographs are representative of cells from at least five different experiments.
Because deletion of the death domain allowed the truncated TNFR1 to translocate to the cell surface, we wondered whether addition of the death domain sequence to a protein expressed on the cell surface would lead to its retention in the Golgi. To answer this question, we made a chimeric construct, gfpTNFR2+DD, consisting of the full-length TNFR2 with the death domain from TNFR1 added to the C-terminus, followed by GFP. Examination by confocal microscopy of HUVEC transfected with gfpTNFR2+DD shows the protein to be distributed on the cell surface as well as in the Golgi (Fig. 3D). Overall, the distributions of gfpTNFR2 and gfpTNFR2+DD were indistinguishable. These data indicate that the death domain was necessary but not sufficient for Golgi retention.
Functional Consequences of Overexpressed TNF Receptor Molecules
In a final series of experiments, we examined whether expression of exogenous TNF receptors in HUVEC would alter responsiveness to TNF. To perform this analysis, we first determined whether transfection of exogenous receptors influenced the total expression of receptor molecules on the cell surface. By FACS analysis, none of the receptor constructs examined produced a dramatic difference in the level of immunoreactive TNFR1 (Table 2). TNFR2 surface expression was increased by transfection of either TNFR2 or TNFR2+DD, as expected, and to a lesser extent by transfection of TNFR1 or TNFR1−DD. However, the changes observed were not large enough to account for any differences in response to ligand.
TNF signaling can lead to either cell activation or cell death. Ordinarily, HUVEC are activated by TNF and are resistant to TNF-induced apoptosis. We examined whether the TNFR constructs induced cell death or sensitized the cells to killing by TNF. HUVEC transfected with the TNFR constructs or empty vector were mock-treated or treated with increasing concentrations of TNF for twelve hours, then fixed and counted. There was no increase in cell death among the HUVEC transfected with the different TNFR constructs, indicating that the TNFR constructs themselves did not kill the cells. Similarly, none of the receptor constructs seemed to sensitize the cells to death by TNF (not shown).
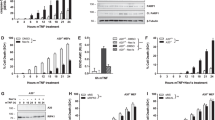
Because binding of TNF to its receptors signals by inducing a clustering of receptor intracellular domains that then interact with adapter proteins leading to the activation of NF-κB and other downstream signals, spontaneous receptor clustering could initiate signaling in a ligand-independent manner (Boldin et al, 1995). To determine whether the TNFR constructs were able to induce ligand-independent signaling, we cotransfected the HUVEC with increasing amounts of each of the TNFR constructs, a κB-luciferase reporter gene and a β-actin Renilla luciferase construct; NF-κB activation was measured by a luciferase assay and normalized to β-actin Renilla luciferase activity to control for transfection efficiency. Western blot analysis verified that the transfected protein levels increased with higher amounts of DNA transfected over the range examined (not shown). Transfection with gfpTNFR1−DD, which lacks the death domain region and is thus unable to interact with the adaptor protein TRADD, did not activate NF-κB activity (Fig. 4). In contrast, transfection with gfpTNFR1 or gfpTNFR2+DD induced a small, dose-dependent increase in NF-κB activity compared with mock-transfected cells. Overexpressed gfpTNFR2 induced a substantially larger increase in ligand-independent signaling. The enhanced signaling capacity of TNFR2 compared with TNFR2+DD suggests that the death domain may inhibit signaling.
Effect of transfected TNFR molecules on ligand-independent NF-κB activation. HUVEC were cotransfected with the reporter vectors NF-κB luciferase and β-actin–Renilla luciferase, pEGFP control vector, and different amounts of the TNFR constructs gfp-TNFR1, gfp-TNFR−DD, gfp-TNFR2, or gfp-TNFR2+DD. Total DNA amount was kept constant with the addition of pEGFP. Cells were split 24 hours after transfection and were not further treated. Sixty hours after transfection, cells were analyzed for NF-κB activation. Results were normalized for transfection efficiency using the Dual Luciferase assay (see “Materials and Methods”) and fold induction of NF-κB activation over control vector was calculated. The full-length TNFR1 (gfp-TNFR1, filled circles), full-length TNFR2 (gfp-TNFR2, filled squares), and the TNFR2 chimera (gfp-TNFR2+DD, open squares) are able to activate NF-κB in the absence of TNF. The death domain-truncated TNFR1 (gfp-TNFR1−DD, open circles) does not show ligand-independent NF-κB activation. Values shown are averages (mean ± sd) of triplicate wells of one experiment, representative of four similar experiments. TNFR1, TNFR2, and TNFR2+DD showed a significant increase over their controls at 8.1 μg of DNA, with p < 0.05 by ANOVA with Bonferroni correction.
Finally, we examined NF-κB activity in the transfected cells treated with TNF. As reported previously in the murine L929 and A9 cells (Brakebusch et al, 1992; Tartaglia and Goeddel, 1992a), TNFR1 lacking the death domain had a dominant negative effect on TNF signaling. Surprisingly, both gfpTNFR1 and gfpTNFR2+DD also inhibited the response to TNF. This inhibitory effect was dose-dependent and consistent with the reduced capacity of gfpTNFR2+DD to activate ligand-independent signaling compared with TNFR2, as described above. In contrast, gfpTNFR2 did not influence the TNF response, neither sensitizing nor blunting the TNF response (Fig. 5). The effects were the same for HUVEC transfected with the constructs lacking the GFP tag, indicating that this dominant negative effect was not due to hindrance from the GFP fusion protein (not shown). These effects were also specific for TNF because none of the constructs inhibited or increased the activation of the same reporter gene by lipopolysaccharide (LPS) (not shown).
Effect of transfected TNFR molecules on TNF-induced NF-κB activation. HUVEC were cotransfected with the reporter vectors NF-κB luciferase and β-actin-Renilla-luciferase, pEGFP control vector and different amounts of the TNFR constructs gfp-TNFR1 (A), gfp-TNFR2 (B), gfp-TNFR1−DD (C), and gfp-TNFR2+DD (D). Total DNA amount was kept constant with the addition of pEGFP. Cells were split 24 hours after transfection, rested for another 24 hours, and either left untreated or treated with increasing concentrations of TNF (0.1, 1.0, and 10 ng/ml) for 12 hours, then harvested and analyzed for NF-κB activation. Results were normalized for transfection efficiency using the Dual Luciferase assay (see “Materials and Methods”). Full-length TNFR1 and the chimera TNFR2+DD, both of which contain the TNFR1 death domain sequence, had a dose-dependent inhibitory effect on TNF signaling of NF-κB activation, despite inducing ligand-independent NF-κB activation. Full-length TNFR2, which also induced ligand-independent NF-κB activation, had no net effect on TNF signaling. In contrast, the death domain-truncated TNFR1−DD showed a dose dependent, dominant negative effect on TNF signaling and did not cause ligand independent NF-κB activation. Each panel is constructed from independent experiments. The values shown are averages (mean ± sd) of triplicate wells of one experiment, representative of four experiments using the same constructs with similar results. Statistical significance was calculated for the 10 ng/ml of TNF dose for each construct versus its control. * p < 0.05 by ANOVA with Bonferroni correction.
Discussion
TNF-activation of EC is an important part of the development of inflammatory processes. These responses are mediated by TNF receptors although it is not known whether the intracellular localization of receptor expression is important for regulating TNF signaling. Although there has been much work on elucidating the biochemical mechanisms of TNF signaling, factors governing the intracellular localizations of TNF and TNF receptors are still not well understood. EC regulate the cellular distribution of endogenous TNF receptors, with TNFR1 predominantly confined to the Golgi apparatus and TNFR2 on the plasma membrane. In this study we have shown that transfected TNFR1 and TNFR2 recapitulate these patterns of localization in HUVEC. Furthermore, we found that the death domain of TNFR1 is necessary, but not sufficient, for Golgi retention. We also showed that overexpression of full-length TNFR1, but not TNFR2, has an inhibitory effect on TNF signaling in EC. Part of this inhibitory effect is mediated through the death domain because expression of a mutant TNFR2 that contains the death domain of TNFR1 also inhibits ligand-dependent activation of NF-κB.
The consequences for the cell of retaining TNFR1 in the Golgi remain unclear. TNF can induce either cell activation or cell death, but the intracellular localization of TNFR1 apparently does not regulate which of these occurs; TNFR1 is predominantly retained in the Golgi in both U937 cells, which are sensitive to TNF-induced apoptosis, and in EC, which are resistant to TNF-induced apoptosis (Jones et al, 1999). One possibility is that intracellular sequestration of the receptor represents a way to modulate sensitivity to TNF signaling. Bennett et al described a similar situation for the death domain-containing, apoptosis-inducing receptor Fas in vascular smooth muscle cells (Bennett et al, 1998). In these cells, Fas is predominantly Golgi-associated but redistributes to the cell surface upon activation of p53, thereby sensitizing the cells to apoptosis. We have not found Fas to localize to the Golgi in HUVEC (Gaeta ML, unpublished observation). Moreover, conditions that mobilize TNFR1 from its intracellular reservoir to the cell surface have not been identified.
Another possibility is that the Golgi pool of TNFR1 participates in signaling, perhaps interacting with the adaptor protein TRADD, which is also Golgi-associated. However, in ECV304 bladder carcinoma and U937 monocytoid cells, TNFR1 and TRADD reside in different Golgi subcompartments and only plasma membrane TNFR1 molecules are able to recruit TRADD in response to TNF (Jones et al, 1999). Interestingly, in HUVEC, some TNFR1 and TRADD molecules reside in the same mannosidase-positive Golgi subcompartment (Gaeta ML, unpublished observation). Perhaps TNFR1 in the Golgi initiates signals in those cell types that synthesize TNF, ie, where ligand is provided access to the Golgi. Finally, it is possible that intracellular TNFR1 interacts with intracellular proteins different from those identified as interacting with plasma membrane receptor. One such candidate could be SODD (silencer of death domains), which has recently been reported to bind to the intracellular region of TNFR1 in the absence of TNF, thereby preventing receptor self-aggregation and blocking the recruitment of TRADD and down-stream signaling (Jiang et al, 1999). However, in preliminary experiments we did not see co-localization of SODD with TNFR1 in the Golgi (Gaeta ML, unpublished observation). Another such candidate could be caveolin-1. Caveolin-1 is a membrane protein associated with the Golgi apparatus, the plasma membrane, and with caveolae, which are specialized invaginations of the plasma membrane. It is thought that caveolin is involved in molecular transport and organization of signaling complexes (Okamoto et al, 1998). This possibility is under current investigation.
In the present study, deletions of the death domain from TNFR1 abolished Golgi retention and incorporation of the death domain at the C-terminus of TNFR2 did not confer Golgi retention. Other death domain sequences also seem insufficient to confer Golgi retention. For example, although Fas is Golgi-associated in vascular smooth muscle cells, Fas does not localize to the Golgi in HUVEC (Gaeta ML, unpublished observation). DR3, another death domain-containing member of the TNFR superfamily (Chinnaiyan et al, 1996; Kitson et al, 1996), interacts with TRADD to signal both apoptosis and NF-κB activation, and also is not seen in the Golgi in HUVEC (Gaeta ML, unpublished observation).
TNFR activation is normally dependent on ligand-induced receptor trimerization. Truncations of the intracellular domain of TNFR1 have been shown to have a dominant negative effect on TNF signaling in L929 cells, presumably by forming non-functional trimers (Tartaglia and Goeddel, 1992a); here we showed a similar effect for TNFR1−DD in HUVEC. Overexpression of full-length TNFR1 has been reported to confer TNF reactivity in mouse 70Z/3 cells, which lack endogenous receptor (Kruppa et al, 1992). Therefore, it was unexpected to find that full-length TNFR1 was inhibitory rather than activating in HUVEC. The mechanism of inhibition by the full-length construct cannot be attributed to a dominant negative effect. One possible explanation for this observation is that overexpression of TNFR1 leads to receptor self-aggregation and ligand-independent signaling in HUVEC and that constitutive activation of this pathway in HUVEC, but not in 70Z/3 cells, may autoregulate the signaling pathway, desensitizing the cells to TNF-induced activation. The inhibitory effect of TNFR1, which was shared by TNFR2+DD, was specific for TNF and did not affect LPS-activation of the κB reporter gene. Homologous desensitization of signaling pathways have been previously observed for activation of c-Jun N-terminal kinase in HUVEC, although it has not been shown for NF-κB (Karmann et al, 1996). This difference may reflect the mechanism of desensitization caused by ligand versus that caused by receptor overexpression. This inhibitory effect of overexpressed TNFR1 also contrasts with the sensitization to TNF-mediated apoptosis observed in other cell lines transfected with the receptor (Boldin et al, 1995; Brakebusch et al, 1992); however, HUVEC are generally resistant to TNF-induced apoptosis in the absence of a second signal (eg, actinomycin or ceramide) and therefore the pathways leading to TNF-induced apoptosis may differ from those leading to NF-κB examined in this study.
The role of TNFR2 in EC remains unclear. Previous studies in EC using monoclonal antibodies and receptor-specific TNF muteins have shown that TNFR2 cannot directly activate EC but facilitates TNF signaling through TNFR1 at low TNF concentrations (Mackay et al, 1993; Slowik et al, 1993). This effect has been explained by ligand passing. In this study, however, overexpression of TNFR2 increased NF-κB activation, but neither enhanced nor inhibited the response to TNF. In contrast, TNFR2+DD had an inhibitory effect, like that of full-length TNFR1, indicating a role for the death domain in mediating this response. Because the transfectants did overexpress TNFR2 on the cell surface, these data are consistent with the interpretation that TNFR2 is normally expressed in HUVEC at a level that is insufficient to mediate ligand-independent signaling, but that the basal level of expression is sufficient to optimally interact with TNFR1, eg, through ligand passing. This issue will require further examination.
In summary, we have identified the death domain of TNFR1 as responsible for mediating Golgi retention of the receptor in HUVEC, and although the death domain is insufficient to cause Golgi retention on its own (eg, when expressed on TNFR2), its overexpression is sufficient to desensitize HUVEC to TNF-mediated NF-κB activation.
Materials and Methods
Antibodies and Reagents
The murine IgG1 monoclonal antibodies (mAb) used in this study were: M2 (anti-FLAG; Kodak IBI, New Haven, Connecticut), anti-TNFR1 and anti-TNFR2 (Genzyme, Cambridge, Massachusetts), a second anti-TNFR1 antibody (from Dr. D. Goeddel, Tularik Inc., South San Francisco, California), K16/16 (a non-binding mAb) (from Dr. D. Mendrick, Brigham and Women's Hospital, Boston, Massachusetts), and anti-GFP (Clontech, Palo Alto, California). The Golgi-specific rabbit polyclonal antibody MannII (rabbit anti-serum against rat liver mannosidase) was a gift from Dr. K. Moremen (University of Georgia, Athens, Georgia). Normal donkey serum (Jackson ImmunoResearch, West Grove, Pennsylvania) and normal goat serum (Sigma Chemical Company, St. Louis, Missouri) were used to reduce non-specific binding.
Secondary antibodies used for immunoreactivity and Western blotting were phycoerythrin-conjugated F(ab′)2 fragment of goat-anti mouse IgG, Texas Red-conjugated donkey anti-mouse IgG, FITC-conjugated donkey anti-rabbit IgG, and horseradish peroxidase-conjugated donkey anti-mouse and donkey anti-rabbit IgG (all from Jackson ImmunoResearch).
Other reagents used were saponin, Triton X-100, paraformaldehyde, and bovine serum albumin (all from Sigma), Collagenase Type I (Worthington Biochemical Company, Freehold, New Jersey), fibronectin (isolated from human plasma by gelatin-Sepharose chromatography), LPS serotype 0111:B4 (Sigma), and recombinant human TNF (Peprotech, Rocky Hill, New Jersey).
Cell Culture
Primary HUVEC were isolated from 3 to 5 umbilical veins, pooled, and cultured as previously described (Gimbrone, 1976) on gelatin-coated culture plastic in medium M199 supplemented with 20% fetal calf serum, 200 μm l-glutamine, 100 U/ml of penicillin and 100 μg/ml of streptomycin (all from Life Technologies, Grand Island, New York), 50 μg/ml of EC growth factor (Collaborative Biomedical Products, Bedford, Massachusetts) and 100 μg/ml of porcine intestinal heparin (Sigma). Cells were passaged by trypsin resuspension and transfected at subculture three.
DNA Constructs and Transfection of HUVEC
A schematic of the expression constructs used in this study is shown in Figure 6. The N-terminal FLAG epitope-tagged full-length TNFR1 (FLAG-TNFR1) and TNFR2 (FLAG-TNFR2) constructs were generated by cloning sequences encoding the receptors (a gift from Immunex, Seattle, Washington) into the FLAG expression vector pRCX3 (a gift from Dr. P. Yurchenco, Robert Wood Johnson Medical School, Piscataway, New Jersey). The deletion constructs FLAG-TNFR1−IC, lacking the intracellular domain (residues 206–426), and FLAG-TNFR1−DD, truncating the C-terminal half of the intracellular domain, which comprise the death domain (residues 315–426), were generated by PCR cloning from FLAG-TNFR1 with a proof-reading DNA polymerase (Vent, New England BioLabs, Beverly, Massachusetts). The GFP expression vector pGL (Invitrogen, Carlsbad, California) was used in cotransfections with the FLAG epitope-tagged constructs for fluorescent flow cytometry. The C-terminal–enhanced GFP fusion constructs gfpTNFR1, gfpTNFR1−DD, and gfpTNFR2 were generated in the pEGFP-N2 expression vector (Clontech) by PCR cloning from the FLAG-tagged TNFR constructs. The chimeric construct gfpTNFR2+DD, consisting of the full-length TNFR2 and the C-terminal portion of the intracellular domain of TNFR1 (residues 315–426), was generated by overlapping PCR cloning from the two full-length receptors. All constructs were confirmed by sequencing. The NF-κB–dependent reporter gene 2κB-firefly luciferase (a gift from Dr. S. Ghosh, Yale University, New Haven, Connecticut) (Min et al, 1996), was used to determine NF-κB activity. In these experiments, a constitutively active, cytokine-independent reporter gene, β-actin–Renilla luciferase (a gift from Dr. S. Ghosh), was used to normalize transfection efficiency.
Schematic representation of DNA constructs. Full-length cDNA for TNFR1 or TNFR2 were cloned into the expression vector pRCX3 downstream of FLAG. The deletion mutants TNFR1−IC and TNFR1−DD were made by inserting stop codons at position 206, truncating the IC region one amino acid past the transmembrane domain, and at position 315, truncating the C-terminal half of the IC region containing the death domain. The GFP fusion constructs were made by inserting the TNFR cDNA into the pEGFP vector. The chimera TNFR2+DD was generated by overlapping PCR of TNFR2 and the C-terminal half of the IC region of TNFR1.
HUVEC grown to 70% confluence on 100 mm or 35 mm diameter plastic culture plates were transfected approximately 18 hours after passage using a modified DEAE-dextran protocol as previously described (Karmann et al, 1996). Transfection efficiencies typically were between 10% and 25%. Cells were cotransfected with pGL plus FLAG-TNFR1, FLAG-TNFR2, or pRCX3; with pEGFP, gfpTNFR1, gfpTNFR1−DD, gfpTNFR2, or gfpTNFR2+DD; or with the κB-firefly luciferase and β-actin–Renilla luciferase reporter vectors plus one of the receptor constructs. Total DNA amount was kept constant by the addition of control vectors. Transfected HUVEC were analyzed 60 hours after transfection.
FACS Analysis of TNFR1 and TNFR2
To quantify surface and total expression of TNFR1 or TNFR2, HUVEC were harvested by collagenase treatment 60 hours after transfection, washed once in serum-containing medium and twice in wash buffer (WB, consisting of filtered 1% BSA in PBS). Cells were kept on ice. Permeabilized cells were fixed with 4% paraformaldehyde for 20 minutes, washed three times with WB, and once with permeabilization buffer (PB, 0.1% saponin, 1% filtered BSA in PBS). Cells were then incubated for 1 hour with primary antibody (10 μg/ml of M2, anti-R1, or anti-R2; K16/16 diluted 1:200) in either WB or PB, followed by three washes. Cells were then incubated for 30 minutes with goat anti-mouse phycoerythrin-labeled antibody diluted 1:50 in either WB or PB. Permeabilized cells were then washed twice with PB, twice with WB, once with PBS, and resuspended with 0.4 ml of PBS. Non-permeabilized cells were washed twice with WB, once with PBS, and resuspended and fixed with 4% paraformaldehyde. Cells were analyzed using a FACSort flow cytometer (Becton Dickinson, San Jose, California) running LYSIS II software. Corrected mean fluorescence intensities were calculated by subtracting the mean fluorescence intensity with irrelevant nonbinding antibody from the mean fluorescence intensity with the specific antibody.
Immunofluorescence and Confocal Microscopy
At 48 hours post-transfection, cells were harvested and replated on fibronectin-coated coverslips, incubated overnight, and immunohistochemisty was performed as previously described (Bradley et al, 1995). Cells were examined using a Nikon Microphot-FXA microscope and a Zeiss LSM 510 confocal microscope. Cells transfected with GFP fusion constructs were plated on fibronectin-coated, 35 mm glass-bottom culture plates (MatTek, Ashland, Massachusetts) and examined live by confocal microscopy. All pictures were taken of cells using the 40X objective.
Reporter Assay for NF-κB Activation
HUVEC were cotransfected with the TNFR constructs and the reporter genes 2κB-firefly luciferase and β-actin–Renilla luciferase. At 24 hours post-transfection the cells were replated onto gelatin-coated 24-well plastic culture plates and incubated for 24 hours. Cells were then treated with TNF (0.1, 1, or 10 ng/ml) or LPS (100 ng/ml). After 12 hours of incubation, cells were assayed for firefly and Renilla luciferase activity using the Dual Luciferase kit (Promega, Madison, Wisconsin).
Western Blotting
Transfected HUVEC were lysed in buffer containing 20 mm Tris-HCl, pH 8, 137 mm NaCl, 2 mm EDTA, 10% glycerol, 1% Triton X-100, 1 mm phenyl-methylsulfonyl fluoride, and the protease inhibitor cocktail Complete, Mini (Boehringer Mannheim, Indianapolis, Indiana), cleared by centrifugation, and electrophoresed in an 8% SDS-PAGE under reducing conditions. The resolved proteins were transferred to Immobilon membranes (Millipore Corporation, Bedford, Massachusetts), immunoreacted with antibodies to GFP, FLAG, TNFR1, or TNFR2 followed by horseradish peroxidase-conjugated secondary antibodies. Reactive proteins were visualized by enhanced chemiluminescence according to the manufacturer (Pierce, Rockford, Illinois).
Statistical Analyses
Statistical analyses were performed using the program Primer of Biostatistics version 3.01. The data in Table 2 were analyzed by Student's paired t test. The data in Figures 4 and 5 were analyzed by ANOVA with Bonferroni correction.
References
Adam D, Wiegmann K, Adam-Klages S, Ruff A, and Kronke M (1996). A novel cytoplasmic domain of the p55 tumor necrosis factor receptor initiates the neutral sphingomyelinase pathway. J Biol Chem 271:14617–14622.
Bazzoni F and Beutler B (1996). The tumor necrosis factor ligand and receptor families. N Engl J Med 334:1717–1725.
Bennett M, Macdonald K, Chan SW, Luzio JP, Simari R, and Weissberg P (1998). Cell surface trafficking of Fas: A rapid mechanism of p53-mediated apoptosis. [See comments, Science 284:1431]. Science 282:290–293.
Bevilacqua MP, Pober JS, Mendrick DL, Cotran RS, and Gimbrone MA Jr (1987). Identification of an inducible endothelial-leukocyte adhesion molecule. Proc Natl Acad Sci U S A 84:9238–9242.
Boldin MP, Mett IL, Varfolomeev EE, Chumakov I, Shemer-Avni Y, Camonis JH, and Wallach D (1995). Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J Biol Chem 270:387–391.
Bradley JR, Thiru S, and Pober JS (1995). Disparate localization of 55-kd and 75-kd tumor necrosis factor receptors in human endothelial cells. Am J Pathol 146:27–32.
Brakebusch C, Nophar Y, Kemper O, Engelmann H, and Wallach D (1992). Cytoplasmic truncation of the p55 tumour necrosis factor (TNF) receptor abolishes signalling, but not induced shedding of the receptor. Embo J 11:943–950.
Carlos TM, Schwartz BR, Kovach NL, Yee E, Rosa M, Osborn L, Chi-Rosso G, Newman B, Lobb R, and Harlan JM (1990). Vascular cell adhesion molecule-1 mediates lymphocyte adherence to cytokine-activated cultured human endothelial cells. Blood 76:965–970.
Chinnaiyan AM, O'Rourke K, Yu GL, Lyons RH, Garg M, Duan DR, Xing L, Gentz R, Ni J, and Dixit VM (1996). Signal transduction by DR3, a death domain-containing receptor related to TNFR-1 and CD95. Science 274:990–992.
Cleveland JL and Ihle JN (1995). Contenders in FasL/TNF death signaling. Cell 81:479–482.
Feinstein E, Kimchi A, Wallach D, Boldin M, and Varfolomeev E (1995). The death domain: A module shared by proteins with diverse cellular functions (letter). Trends Biochem Sci 20:342–344.
Gimbrone MA Jr (1976). Culture of vascular endothelium. Prog Hemost Thromb 3:1–28.
Haridas V, Darnay BG, Natarajan K, Heller R, and Aggarwal BB (1998). Overexpression of the p80 TNF receptor leads to TNF-dependent apoptosis, nuclear factor-kappa B activation, and c-Jun kinase activation. J Immunol 160:3152–3162.
Hsu H, Huang J, Shu HB, Baichwal V, and Goeddel DV (1996a). TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity 4:387–396.
Hsu H, Shu HB, Pan MG, and Goeddel DV (1996b). TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299–308.
Hsu H, Xiong J, and Goeddel DV (1995). The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell 81:495–504.
Jiang Y, Woronicz JD, Liu W, and Goeddel DV (1999). Prevention of constitutive TNF receptor 1 signaling by silencer of death domains [published erratum appears in Science 283:1852]. Science 283:543–546.
Jones SJ, Ledgerwood EC, Prins JB, Galbraith J, Johnson DR, Pober JS, and Bradley JR (1999). TNF recruits TRADD to the plasma membrane but not the trans-Golgi network, the principal subcellular location of TNF-R1. J Immunol 162:1042–1048.
Karmann K, Min W, Fanslow WC, and Pober JS (1996). Activation and homologous desensitization of human endothelial cells by CD40 ligand, tumor necrosis factor, and interleukin 1. J Exp Med 184:173–182.
Kitson J, Raven T, Jiang YP, Goeddel DV, Giles KM, Pun KT, Grinham CJ, Brown R, and Farrow SN (1996). A death-domain-containing receptor that mediates apoptosis. Nature 384:372–375.
Kruppa G, Thoma B, Machleidt T, Wiegmann K, and Kronke M (1992). Inhibition of tumor necrosis factor (TNF)-mediated NF-kappa B activation by selective blockade of the human 55-kDa TNF receptor. J Immunol 148:3152–3157.
Machleidt T, Kramer B, Adam D, Neumann B, Schutze S, Wiegmann K, and Kronke M (1996). Function of the p55 tumor necrosis factor receptor “death domain” mediated by phosphatidylcholine-specific phospholipase C. J Exp Med 184:725–733.
Mackay F, Loetscher H, Stueber D, Gehr G, and Lesslauer W (1993). Tumor necrosis factor alpha (TNF-alpha)-induced cell adhesion to human endothelial cells is under dominant control of one TNF receptor type, TNF-R55. J Exp Med 177:1277–1286.
Min W, Ghosh S, and Lengyel P (1996). The interferon-inducible p202 protein as a modulator of transcription: Inhibition of NF-kappa B, c-Fos, and c-Jun activities. Mol Cell Biol 16:359–368.
Okamoto T, Schlegel A, Scherer PE, and Lisanti MP (1998). Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem 273:5419–5422.
Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi-Rosso G, and Lobb R (1989). Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell 59:1203–1211.
Park A and Baichwal VR (1996). Systematic mutational analysis of the death domain of the tumor necrosis factor receptor 1-associated protein TRADD. J Biol Chem 271:9858–9862.
Park YC, Burkitt V, Villa AR, Tong L, and Wu H (1999). Structural basis for self-association and receptor recognition of human TRAF2. Nature 398:533–538.
Pober JS, Bevilacqua MP, Mendrick DL, Lapierre LA, Fiers W, and Gimbrone MA Jr (1986). Two distinct monokines, interleukin 1 and tumor necrosis factor, each independently induce biosynthesis and transient expression of the same antigen on the surface of cultured human vascular endothelial cells. J Immunol 136:1680–1687.
Pober JS and Cotran RS (1990). Cytokines and endothelial cell biology. Physiol Rev 70:427–451.
Pober JS, Lapierre LA, Stolpen AH, Brock TA, Springer TA, Fiers W, Bevilacqua MP, Mendrick DL, and Gimbrone MA Jr (1987). Activation of cultured human endothelial cells by recombinant lymphotoxin: Comparison with tumor necrosis factor and interleukin 1 species. J Immunol 138:3319–3324.
Rice GE and Bevilacqua MP (1989). An inducible endothelial cell surface glycoprotein mediates melanoma adhesion. Science 246:1303–1306.
Slowik MR, De Luca LG, Fiers W, and Pober JS (1993). Tumor necrosis factor activates human endothelial cells through the p55 tumor necrosis factor receptor but the p75 receptor contributes to activation at low tumor necrosis factor concentration. Am J Pathol 143:1724–1730.
Smith CA, Farrah T, and Goodwin RG (1994). The TNF receptor superfamily of cellular and viral proteins: Activation, costimulation, and death. Cell 76:959–962.
Strieter RM, Kunkel SL, Showell HJ, and Marks RM (1988). Monokine-induced gene expression of a human endothelial cell-derived neutrophil chemotactic factor. Biochem Biophys Res Commun 156:1340–1345.
Strieter RM, Wiggins R, Phan SH, Wharram BL, Showell HJ, Remick DG, Chensue SW, and Kunkel SL (1989). Monocyte chemotactic protein gene expression by cytokine-treated human fibroblasts and endothelial cells. Biochem Biophys Res Commun 162:694–700.
Tartaglia LA, Ayres M, Wong GH, and Goeddel DV (1993). A novel domain within the 55 kd TNF receptor signals cell death. Cell 74:845–853.
Tartaglia LA and Goeddel DV (1992a). Tumor necrosis factor receptor signaling. A dominant negative mutation suppresses the activation of the 55-kDa tumor necrosis factor receptor. J Biol Chem 267:4304–4307.
Tartaglia LA and Goeddel DV (1992b). Two TNF receptors. Immunol Today 13:151–153.
Acknowledgements
We thank Ms. Louise Camera Benson and Ms. Gwendoline Davis for assistance in cell culture, Mr. Barry Mook for assistance in preparing the mutation constructs, Mr. Roger Babbitt and Dr. Rocio Sierra-Honigmann for help with microscopy, and Drs. Werner Lesslauer, Mae Gailani, Keyvan Mahboubi, Lisa Madge, and Xiao Feng for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by grants from the Pediatric Scientist Development Program and the National Institutes of Health (HL-36003).
Rights and permissions
About this article
Cite this article
Gaeta, M., Johnson, D., Kluger, M. et al. The Death Domain of Tumor Necrosis Factor Receptor 1 Is Necessary but Not Sufficient for Golgi Retention of the Receptor and Mediates Receptor Desensitization. Lab Invest 80, 1185–1194 (2000). https://doi.org/10.1038/labinvest.3780126
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.3780126
This article is cited by
-
A TNFR1–UBCH10 axis drives lung squamous cell carcinoma dedifferentiation and metastasis through a cell-autonomous signaling loop
Cell Death & Disease (2022)
-
Intracellular application of TNF-alpha impairs cell to cell communication via gap junctions in glioma cells
Journal of Neuro-Oncology (2008)
-
Increased ICAM-1 Expression Causes Endothelial Cell Leakiness, Cytoskeletal Reorganization and Junctional Alterations
Journal of Investigative Dermatology (2007)