Abstract
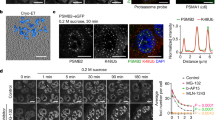
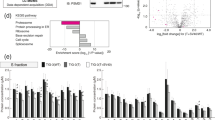
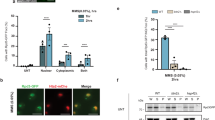
The p53 tumor suppressor is a nucleocytoplasmic shuttling protein that is found predominantly in the nucleus of cells. In addition to mutation, abnormal p53 cellular localization is one of the mechanisms that inactivate p53 function. To further understand features of p53 that contribute to the regulation of its trafficking within the cell, we analysed the subnuclear localization of wild-type and mutant p53 in human cells that were either permeabilized with detergent or treated with the proteasome inhibitor MG132. We, here, show that either endogenously expressed or exogenously added p53 protein localizes to the nucleolus in detergent-permeabilized cells in a concentration- and ATP hydrolysis-dependent manner. Two discrete regions within the carboxyl terminus of p53 are essential for nucleolar localization in permeabilized cells. Similarly, localization of p53 to the nucleolus after proteasome inhibition in unpermeabilized cells requires sequences within the carboxyl terminus of p53. Interestingly, genotoxic stress markedly decreases the association of p53 with the nucleolus, and phosphorylation of p53 at S392, a site that is modified by such stress, partially impairs its nucleolar localization. The possible significance of these findings is discussed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Appella E, Anderson CW . (2001). Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772.
Ashcroft M, Taya Y, Vousden KH . (2000). Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol 20: 3224–3233.
Benninghoff J, Kartarius S, Teleb Z, Selter H, Unteregger G, Zwergel T et al. (1999). Two different forms of p53 localized differently within cells of urogenital tumours. Cancer Lett 144: 55–64.
Budde A, Grummt I . (1999). p53 represses ribosomal gene transcription. Oncogene 18: 1119–1124.
Bykov VJ, Issaeva N, Selivanova G, Wiman KG . (2002a). Mutant p53-dependent growth suppression distinguishes PRIMA-1 from known anticancer drugs: a statistical analysis of information in the National Cancer Institute database. Carcinogenesis 23: 2011–2018.
Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P et al. (2002b). Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med 8: 282–288.
Carmo-Fonseca M, Mendes-Soares L, Campos I . (2000). To be or not to be in the nucleolus. Nat Cell Biol 2: E107–E112.
Carmo-Fonseca M, Platani M, Swedlow JR . (2002). Macromolecular mobility inside the cell nucleus. Trends Cell Biol 12: 491–495.
Chan PK, Qi Y, Amley J, Koller CA . (1996). Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett 100: 191–197.
Chen X, Ko LJ, Jayaraman L, Prives C . (1996). p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev 10: 2438–2451.
Colombo E, Marine JC, Danovi D, Falini B, Pelicci PG . (2002). Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol 4: 529–533.
Daniely Y, Dimitrova DD, Borowiec JA . (2002). Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol Cell Biol 22: 6014–6022.
Dundr M, Leno GH, Hammarskjold ML, Rekosh D, Helga-Maria C, Olson MO . (1995). The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev protein. J Cell Sci 108 (Part 8): 2811–2823.
Fernandez-Fernandez MR, Veprintsev DB, Fersht AR . (2005). Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc Natl Acad Sci USA 102: 4735–4740.
Ghisolfi-Nieto L, Joseph G, Puvion-Dutilleul F, Amalric F, Bouvet P . (1996). Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol 260: 34–53.
Giaccia AJ, Kastan MB . (1998). The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12: 2973–2983.
Gobert C, Bracco L, Rossi F, Olivier M, Tazi J, Lavelle F et al. (1996). Modulation of DNA topoisomerase I activity by p53. Biochemistry 35: 5778–5786.
Haaf T, Ward DC . (1996). Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res 224: 163–173.
Horky M, Wurzer G, Kotala V, Anton M, Vojtesek B, Vacha J et al. (2001). Segregation of nucleolar components coincides with caspase-3 activation in cisplatin-treated HeLa cells. J Cell Sci 114: 663–670.
Issaeva N, Friedler A, Bozko P, Wiman KG, Fersht AR, Selivanova G . (2003). Rescue of mutants of the tumor suppressor p53 in cancer cells by a designed peptide. Proc Natl Acad Sci USA 100: 13303–13307.
Jayaraman J, Prives C . (1995). Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell 81: 1021–1029.
Jayaraman L, Freulich E, Prives C . (1997). Functional dissection of p53 tumor suppressor protein. Methods Enzymol 283: 245–256.
Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C et al. (1991). Identification of p53 as a sequence-specific DNA-binding protein. Science 252: 1708–1711.
Klibanov SA, O’Hagan HM, Ljungman M . (2001). Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J Cell Sci 114: 1867–1873.
Lamond AI, Sleeman JE . (2003). Nuclear substructure and dynamics. Curr Biol 13: R825–R825.
Latonen L, Kurki S, Pitkanen K, Laiho M . (2003). p53 and MDM2 are regulated by PI-3-kinases on multiple levels under stress induced by UV radiation and proteasome dysfunction. Cell Signal 15: 95–102.
Leung AK, Andersen JS, Mann M, Lamond AI . (2003). Bioinformatic analysis of the nucleolus. Biochem J 376: 553–569.
Leung AK, Lamond AI . (2003). The dynamics of the nucleolus. Crit Rev Eukaryot Gene Expr 13: 39–54.
Liang SH, Clarke MF . (2001). Regulation of p53 localization. Eur J Biochem 268: 2779–2783.
Liu G, Chen X . (2002). The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene 21: 7195–7204.
Llanos S, Clark PA, Rowe J, Peters G . (2001). Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol 3: 445–452.
Marciniak RA, Lombard DB, Johnson FB, Guarente L . (1998). Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci USA 95: 6887–6892.
Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ . (1994). The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol 14: 7414–7420.
Mayer C, Grummt I . (2005). Cellular stress and nucleolar function. Cell Cycle 4: 1036–1038.
Menendez D, Inga A, Resnick MA . (2006). The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol Cell Biol 26: 2297–2308.
Mogaki M, Hirota M, Chaney WG, Pour PM . (1993). Comparison of p53 protein expression and cellular localization in human and hamster pancreatic cancer cell lines. Carcinogenesis 14: 2589–2594.
Olson MO, Dundr M . (2005). The moving parts of the nucleolus. Histochem Cell Biol 123: 203–216.
Olson MO, Dundr M, Szebeni A . (2000). The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol 10: 189–196.
Pokrovskaja K, Mattsson K, Kashuba E, Klein G, Szekely L . (2001). Proteasome inhibitor induces nucleolar translocation of Epstein–Barr virus-encoded EBNA-5. J Gen Virol 82: 345–358.
Poyurovsky MV, Jacq X, Ma C, Karni-Schmidt O, Parker PJ, Chalfie M et al. (2003). Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell 12: 875–887.
Prives C, Hall PA . (1999). The p53 pathway. J Pathol 187: 112–126.
Prives C, Manley JL . (2001). Why is p53 acetylated? Cell 107: 815–818.
Resnitzky D, Gossen M, Bujard H, Reed SI . (1994). Acceleration of the G1/S phase transition by expression of cyclins D1 and E with an inducible system. Mol Cell Biol 14: 1669–1679.
Rokaeus N, Klein G, Wiman KG, Szekely L, Mattsson K . (2006). PRIMA-1(MET) induces nucleolar accumulation of mutant p53 and PML nuclear body-associated proteins. Oncogene [Epub ahead of print].
Rubbi CP, Milner J . (2000). Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19: 85–96.
Shirangi TR, Zaika A, Moll UM . (2002). Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J 16: 420–422.
Stros M, Reich J . (1998). Formation of large nucleoprotein complexes upon binding of the high-mobility-group (HMG) box B-domain of HMG1 protein to supercoiled DNA. Eur J Biochem 251: 427–434.
Tao W, Levine AJ . (1999). P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA 96: 6937–6941.
Thomas F, Kutay U . (2003). Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci 116: 2409–2419.
Tsai RY, McKay RD . (2002). A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 16: 2991–3003.
Visintin R, Amon A . (2000). The nucleolus: the magician's hat for cell cycle tricks. Curr Opin Cell Biol 12: 752.
Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A . (1998). The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk- dependent phosphorylation. Mol Cell 2: 709–718.
Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D . (1999). Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1: 20–26.
Wesierska-Gadek J, Schloffer D, Kotala V, Horky M . (2002). Escape of p53 protein from E6-mediated degradation in HeLa cells after cisplatin therapy. Int J Cancer 101: 128–136.
Wong JM, Kusdra L, Collins K . (2002). Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol 4: 731–736.
Xirodimas DP, Stephen CW, Lane DP . (2001). Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res 270: 66–77.
Young PJ, Day PM, Zhou J, Androphy EJ, Morris GE, Lorson CL . (2002). A direct interaction between the survival motor neuron protein and p53 and its relationship to spinal muscular atrophy. J Biol Chem 277: 2852–2859.
Zatsepina OV, Voronkova LN, Sakharov VN, Chentsov YS . (1989). Ultrastructural changes in nucleoli and fibrillar centers under the effect of local ultraviolet microbeam irradiation of interphase culture cells. Exp Cell Res 181: 94–104.
Zerrahn J, Deppert W, Weidemann D, Patschinsky T, Richards F, Milner J . (1992). Correlation between the conformational phenotype of p53 and its subcellular location. Oncogene 7: 1371–1381.
Zhang Y, Xiong Y, Yarbrough WG . (1998). ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734.
Zimber A, Nguyen QD, Gespach C . (2004). Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell Signal 16: 1085–1104.
Zolotukhin AS, Felber BK . (1999). Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol 73: 120–127.
Acknowledgements
We are particularly grateful to V Gottifredi and K De Vos for guidance during the earlier stages of this work. Thanks to J Ahn, N Baptiste, JC Bulinsky, M Poyurovsky, RPT Tanaka and M Urist for advice and help and to E Freulich who provided expert technical assistance. This work was supported by NIH Grants CA58316 and CA87497.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
Rights and permissions
About this article
Cite this article
Karni-Schmidt, O., Friedler, A., Zupnick, A. et al. Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene 26, 3878–3891 (2007). https://doi.org/10.1038/sj.onc.1210162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1210162
Keywords
This article is cited by
-
Dual regulation of p53 by the ribosome maturation factor SBDS
Cell Death & Disease (2020)
-
Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability
Oncogene (2011)
-
p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis
Oncogene (2008)
-
A high-content chemical screen identifies ellipticine as a modulator of p53 nuclear localization
Apoptosis (2008)