Abstract
This study aimed to explore the molecular epidemiology characteristics of deafness susceptibility genes in neonates in northern Guangdong and provide a scientific basis for deafness prevention and control. A total of 10,183 neonates were recruited between January 2018 and December 2022 at Yuebei People's Hospital. Among these, a PCR hybridization screening group of 8276 neonates was tested for four deafness genes: GJB2, SLC26A4, mtDNA, and GJB3 by PCR hybridization. Another group used next-generation sequencing (NGS) to detect genetic susceptibility genes in 1907 neonates. In PCR hybridization screening group, 346 (4.18%) of 8276 neonates were found to be carriers of the deafness gene. Among these, 182 (2.2%) had GJB2 variants, 114 (1.38%) had SLC26A4 variants, 35 (0.42%) had mtDNA variants, and 15 (0.18%) had GJB3 variants. In NGS Screening Group, 195 out of 1907 neonates were found to be carriers of the deafness gene, with a positive rate of 10.22%. Among these, 137 (7.18%) had GJB2 variants, 41 (2.15%) had SLC26A4 variants, 11 (0.58%) had mtDNA variants, and 6 (0.31%) had GJB3 variants. The prevalence of deafness gene variants was high in Northern Guangdong Province. The most common gene for deafness was GJB2, followed by SLC26A4 and mtDNA. GJB3 variants are rare. Compared with PCR hybridization method, NGS technology can expand the screening scope and greatly improve the detection rate of deafness genes. The c.109G>A of GJB2 was found to occur at a high frequency, which should be considered. Therefore, it is important to conduct neonatal deafness gene screening to prevent and control hereditary deafness.
Similar content being viewed by others
Introduction
According to the World Health Organization (WHO), approximately 500 million people worldwide suffer from disabling deafness. Deafness is the fourth leading cause of disability, accounting for 5.8% of all causes1. Ouyang XM reported in 2009 that China's population is approximately 1.3 billion. An estimated 30,000 infants are born with congenital sensorineural hearing loss each year2. Approximately 70% of congenital hearing loss cases are estimated to have genetic causes3. The deafness genes carried out by our population mainly included GJB2 (DFNB1A, MIM 220,290), SLC26A4 (PDS, MIM 274,600), mitochondrial DNA (mtDNA), (Mitochondrial disease, MIM 561,000), and GJB3 (DFNB1A, MIM 220,290)4. There are regional, ethnic, and methodological differences between deafness genes5. Currently, PCR hybridization method is used to screen for deafness genes in the clinic and only 9–13 variant sites of these four genes are detected6. In this study, we used PCR hybridization and next-generation sequencing (NGS) methods to detect deafness susceptibility genes, explore the molecular epidemiological characteristics of deafness genes in neonates in northern Guangdong province, and provide a scientific basis for the accurate prevention and treatment of deafness.
Materials and methods
Subjects
A total of 10,183 neonates were enrolled between January 2018 and December 2022 at the Yuebei People's Hospital. Screening of four deafness genes (GJB2, SLC26A4, mtDNA and GJB3) without knowing the result of hearing test. Among them, 8276 cases were detected by PCR hybridization, and 1907 cases were detected by NGS. This study was approved by the Ethics Committee of Yuebei People's Hospital (KY-2021-219). Informed consent was obtained from the neonatal guardian, and an informed consent form was signed. All methods followed relevant guidelines and standard operating procedures (SOPs).
Specimen collection
Dried blood spots from neonates were collected according to the blood collection standards and placed in a sealed bag after natural drying; specimens were sent to the hospital's clinical PCR laboratory department for immediate detection or stored at 2–8 °C.
PCR hybridization screening
Thirteen variant sites in the four deafness genes were detected using the deafness susceptibility gene kit (Guangdong Kaipu Biotechnology Co., Ltd.). The test kit was approved by the National Medical Products Administration (NMPA, no. 20153401698). GJB2 included c.35delG, c.155delTCTG, c.176del16, c.235delC, and c.299delAT. SLC26A4 includes c.1229C>T, c.2168A>G and c.919-2A>G. mtDNA includes m.1555A>G, m.1494C>T, m.7445A>G and m.12201T>C. GJB3 c.538C>T. The laboratory operation and positive judgment standard were based on the kit instructions and SOPs. The PCR amplification system included genomic DNA (2 μL), a PCR reaction solution (27.5 μL), and DNA polymerase (0.5 μL). PCR reaction conditions were as follows: pre-denaturation at 95 °C for 9 min, denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, elongationat 72 °C for 1 min, 40 cycles, at 72°Cfor 5 min, and storage at 4 °C.
NGS screening
The target exons of four deafness susceptibility genes were sequenced using the hereditary deafness genetic test kit (BGI Genomics, China). The test kit has been approved by the National Medical Products Administration (NMPA, no. 20203400432). Within ten bases both ends of the exons were analyzed. The basis for estimating the pathogenicity of gene variants is the joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen)7.
Statistical analysis
Data analysis was performed using the SPSS22.0 software (IBM, USA). Data are presented as case (n) or percentage (%).
Ethics statement
This research was approved by the Ethics Committees of Yuebei People’s Hospital.
Results
Result of PCR hybridization screening group
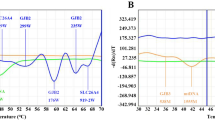
Among 8276 neonates, 346 cases were found to have deafness gene variants, with a positive rate of 4.18%. Among these, 182 (2.2%, 182/8276) had GJB2 variants, 114 (1.38%, 114/8276) had SLC26A4 variants, 35 (0.42%, 35/8276) had mtDNA variants, and 15 (0.18%, 15/8276) had GJB3 variants. The most common deafness gene was GJB2, with c.235delC as its hotspot variant, followed by SLC26A4 and c.919-2A>G, as its hotspot variant (Table 1 and Fig. 1).
Result of NGS screening group
Of the 1907 neonates, 195 were carriers of the deafness gene, with a positivity rate of 10.22%. Among them, 137 (7.18%, 137/1907) had GJB2 variants, 41 (2.15%, 41/1907) had SLC26A4 variants, 11 (0.58%, 11/1907) had mtDNA variants, and 6 (0.31%, 6/1907) had GJB3 variants. The most common deafness gene was GJB2, with c.109G>A as its hotspot variant, followed by SLC26A4, and c.919-2A>G as its hotspot variant (Table 2 and Fig. 2).
Discussion
The detection rate of deafness genes by PCR hybridization screening was only 4.18%, which was lower than the 4.78% of genetic deafness testing results in Chinese newborns8. However, that of NGS screening increased to 10.22%, which improved detection efficiency. Both methods found GJB2 to be the most common deafness gene, followed by SLC26A4 and mtDNA, GJB3 variants were rare, with positive rates of 2.2%, 1.38%, 0.42%, and 0.18%, respectively, consistent with other reports9,10.
GJB2 is the most common variant associated with hereditary deafness11, which encodes the gap junction protein connexin-26 (CX26) on chromosome 13q12.11, which variants and causes congenital sensorineural deafness12. The c.235delC was the most common variant in PCR hybridization screening. Significantly, we found that the c.109G>A variant was the most common in NGS screening. It is possible that the c.109G>A variant was not included in PCR hybridization screening. The pathogenicity of the c.109G>A variant is controversial because of the heterogeneity of the hearing phenotype resulting from the variant at this locus. The c.109G>A homozygous variant, which is considered a polymorphism, can also be detected in individuals with normal hearing. Others may even display severe-to-extreme hearing loss13. The ClinGen hearing loss expert panel determined that c.109G>A is a causative factor for autosomal recessive non-syndromic hearing loss with variable expression and incomplete penetrance14. According to ACMG, the c.109G>A variant is predicted to be a pathogenic variant15. In this study, five patients with the c.109G>A homozygous variant and two with heterozygous variants did not pass hearing screening. Further follow-up and investigation are needed to determine whether their hearing can be restored.
SLC26A4 is the second most mutated gene in hereditary deafness, and c.919-2A>G is a hotspot variant. SLC26A4 is located on 7q22.3 and encodes the pendrin protein, which causes non-syndromic deafness of the vestibular aqueduct to dilate16. Such patients should avoid hard blows, sneezing, blowing nose, head trauma, and other inducements that can effectively prevent deafness17.
MtDNA variants are one of the causes of drug-induced deafness, leading to cochlear and vestibular cell dysfunction18. Using aminoglycosides can lead to deafness. Patients with delayed deafness and their maternal members should be banned from aminoglycoside antibiotics for life19. Therefore, screening for deafness genes before aminoglycoside antibiotics can effectively prevent the tragedy of one-needle deafness.
GJB3 variant is rare, is located on 1p34.3 and encodes gap junction protein 31. Variants cause acquired delayed-onset sensor-neural deafness20. The carrier rate of GJB3 variant is low in China4,9. In this study, the variant rate of GJB3 was also low, at only 0.19%.
Studies have revealed that the results of deafness gene screening can also guide the evaluation of the effectiveness of hearing aids and cochlear implants. For example, patients with deafness associated with GJB2 or SLC26A4 have a good prognosis after cochlear implantation21. The limitation of this study is insufficient information on the subjects, such as sex, twins, hearing loss, and other clinical features. Making it difficult to directly compare the results with previous data.
Conclusions
In brief, the key finding of our investigation pertains to the use of NGS, which surpasses the constraints of conventional screening methods in identifying previously overlooked diagnoses. NGS technology has expanded the scope of deafness gene screening and greatly improved the detection rate. Therefore, it is necessary to include deafness gene testing in hearing screening. High-risk groups can be identified early for intervention or diagnostic guidance. This is significant for individualized and accurate prevention and treatment of deafness.
Data availability
The original data can be obtained by contacting the corresponding author on reasonable request, or available in the China National Center for Bioinformation, [https://bigd.big.ac.cn/gsa-human]. The assigned accession ID is: HRA006031.
References
Wilson, B. S., Tucci, D. L., O’Donoghue, G. M., Merson, M. H. & Frankish, H. A Lancet Commission to address the global burden of hearing loss. Lancet 393, 2106–2108. https://doi.org/10.1016/S0140-6736(19)30484-2 (2019).
Ouyang, X. M. et al. The genetic bases for non-syndromic hearing loss among Chinese. J. Hum. Genet. 54, 131–140. https://doi.org/10.1038/jhg.2009.4 (2009).
Writing Group For Practice Guidelines For Diagnosis And Treatment Of Genetic Diseases Medical Genetics Branch Of Chinese Medical Association, Yuan, H., Dai, P., Liu, Y. & Yang, T. Clinical practice guidelines for hereditary non-syndromic deafness. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 37, 269–276. https://doi.org/10.3760/cma.j.issn.1003-9406.2020.03.008 (2020).
Zhang, J. et al. The Frequency of common deafness-associated variants among 3,555,336 newborns in China and 141,456 individuals across seven populations worldwide. Ear Hear. 44, 232–241. https://doi.org/10.1097/AUD.0000000000001274 (2023).
Souissi, A., Gibriel, A. A. & Masmoudi, S. Genetics and meta-analysis of recessive non-syndromic hearing impairment and Usher syndrome in Maghreb population: Lessons from the past, contemporary actualities and future challenges. Hum. Genet. 141, 583–593. https://doi.org/10.1007/s00439-021-02314-y (2022).
Hao, Z. et al. Large scale newborn deafness genetic screening of 142,417 neonates in Wuhan, China. PLoS ONE. 13, e0195740. https://doi.org/10.1371/journal.pone.0195740 (2018).
Riggs, E. R. et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. 22, 245–257. https://doi.org/10.1038/s41436-019-0686-8 (2020).
Wang, Q. et al. Nationwide population genetic screening improves outcomes of newborn screening for hearing loss in China. Genet. Med. 21, 2231–2238. https://doi.org/10.1038/s41436-019-0481-6 (2019).
Dai, P. et al. Concurrent Hearing and Genetic Screening of 180,469 Neonates with Follow-up in Beijing. China. Am. J. Hum. Genet. 105, 803–812. https://doi.org/10.1016/j.ajhg.2019.09.003 (2019).
Zhu, Y. et al. Association between expanded genomic sequencing combined with hearing screening and detection of hearing loss among newborns in a neonatal intensive care unit. JAMA Netw. Open. 5, e2220986. https://doi.org/10.1001/jamanetworkopen.2022.20986 (2022).
Mahdieh, N., Rabbani, B., Wiley, S., Akbari, M. T. & Zeinali, S. Genetic causes of nonsyndromic hearing loss in Iran in comparison with other populations. J. Hum. Genet. 55, 639–648. https://doi.org/10.1038/jhg.2010.96 (2010).
Mishra, S., Pandey, H., Srivastava, P., Mandal, K. & Phadke, S. R. Connexin 26 (GJB2) mutations associated with non-syndromic hearing loss (NSHL). Indian J. Pediatr. 85, 1061–1066. https://doi.org/10.1007/s12098-018-2654-8 (2018).
Lin, Y. F., Lin, H. C., Tsai, C. L. & Hsu, Y. C. GJB2 mutation spectrum in the Taiwanese population and genotype-phenotype comparisons in patients with hearing loss carrying GJB2 c.109G>A and c.235delC mutations. Hear Res. 413, 108135. https://doi.org/10.1016/j.heares.2020.108135 (2022).
Shen, J. et al. Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen hearing loss expert panel. Genet. Med. 21, 2442–2452. https://doi.org/10.1038/s41436-019-0535-9 (2019).
Jiang, H. et al. A novel compound heterozygous mutation in the GJB2 gene is associated with non-syndromic hearing loss in a Chinese family. Biosci. Trends. 12, 470–475. https://doi.org/10.5582/bst.2018.01156 (2018).
Li, M. et al. Digenic inheritance of mutations in EPHA2 and SLC26A4 in Pendred syndrome. Nat. Commun. 11, 1343. https://doi.org/10.1038/s41467-020-15198-9 (2020).
Danilchenko, V. Y., Zytsar, M. V., Maslova, E. A. & Posukh, O. L. Selection of diagnostically significant regions of the SLC26A4 gene involved in hearing loss. Int. J. Mol. Sci. 23, 13453. https://doi.org/10.3390/ijms232113453 (2022).
Ding, Y., Teng, Y., Guo, Q. & Leng, J. Mitochondrial Trna Gln 4394C>T mutation may contribute to the clinical expression of 1555A>G-induced deafness. Genes (Basel). 13, 1794. https://doi.org/10.3390/genes13101794 (2022).
Yang, T., Guo, L., Wang, L. & Yu, X. Diagnosis, intervention, and prevention of genetic hearing loss. Adv. Exp. Med. Biol. 1130, 73–92. https://doi.org/10.1007/978-981-13-6123-4-5 (2019).
Huang, S. et al. The relationship between the GJB3 c.538C>T variant and hearing phenotype in the Chinese population. Int. J. Pediatr. Otorhinolaryngol. 102, 67–70. https://doi.org/10.1016/j.ijporl.2017.09.001 (2017).
Seligman, K. L. et al. Genetic causes of hearing loss in a large cohort of cochlear implant recipients. Otolaryngol. Head Neck Surg. 166, 734–737. https://doi.org/10.1177/01945998211021308 (2022).
Funding
This work was supported by the Science and Technology Project of Shaoguan (210926144531422), the Health Research Project of Shaoguan (Y22091), and Yuebei People’s Hospital Clinical Scientific Research Project (RS-04).
Author information
Authors and Affiliations
Contributions
Z.M., J.Q., and S.F. conceived and designed the study. Z.M. and W.H. analyzed data and drafted the manuscript and SF revised it. Z.M., Y.L. and M.Y. conducted the laboratory testing. J.X. and S.F. performed the genetic diagnosis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ma, Z., Huang, W., Xu, J. et al. Analysis of deafness susceptibility gene of neonates in northern Guangdong, China. Sci Rep 14, 362 (2024). https://doi.org/10.1038/s41598-023-49530-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-49530-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.